RetrogeneDB ID: | retro_mmus_42 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 4:72851309..72852569(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000059343 | |
| Aliases: | Aldoart1, 4921524E03Rik, Aldo1-ps, Aldo1-ps2, Aldoa-ps2 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Aldoa | ||
| Ensembl ID: | ENSMUSG00000030695 | ||
| Aliases: | Aldoa, Aldo-1, Aldo1 | ||
| Description: | aldolase A, fructose-bisphosphate [Source:MGI Symbol;Acc:MGI:87994] |

Retrocopy-Parental alignment summary:
>retro_mmus_42
ATGGCAACGCACAGGCAAGATGTGTCCATCTTCAACATGACCCGCCTGTCCCTGGCTATGGCTTTTTCCTTTCCCCCAGA
TGCCAATGAGCAACCCCACTCTGGGCTGGACAACACCCACCAACAGACAAAGGAGTTAGGAAAGGAAAGTACTACCACCG
GAACTATGCCCTGCCCATACCCAGCACTGACCACAGAGCAGAAGAAAGAGCTCTCTGACATCGCTCACCGAATTGTGGCT
CCAGGCAAGGGCATCCTGGCTGCAGATGAGTCCATTGGAAGCATGGGCAATCGCCTGCAGTCCATTGGCACTGAGAACAC
TGAAGAGAACAGGCGCTTCTTCCGCCAGTTGCTGCTGACTGCTGATGACCGTGTGAATCCCTGCATTGGGGGTGTGATCC
TCTTCCATGAGACACTGTACGAGAAGGCAGATGATGGACGTCCTTTCCCCCAAGTTATTAAGTCCAAGGGTGGTGTTGTG
GGCATTAAGGTAGATAAGGGTGTGGTGCCCCTGGCAGGAACCAATGGCGAGACTACTACCCAAGGGCTAGATGGGCTGTC
TGAACGCTGTGCCCAGTATAAGAAGGATGGAGCCGACTTTGCCAAGTGGCGCTGTGTGCTAAAGATTGGGAAACATACTC
CCTCGCCCCTTGCCATCATGGAGAATGCCAATGTTCTGGCCCGTTATGCCAGCATCTGCCAGCAGAATGGCATTGTACCC
ATTGTGGAGCCTGAAATTCTCCCTGATGGTGACCATGACTTGAGTTGTTGCCAGTATGTAACTGAGAAGGTACTGGCAGC
TGTCTACAAGGCTCTGAGTGACCACCATGTCTATCTGGAAGGCACATTGTTGAAGCCCAACATGGTCACCCCTGGTCATG
CTTGCACCCAGAAATTTTCCAATGAGGAGATTGCCATGGCAACAGTCACAGCACTTCGTCGCACAGTGCCACCTGCTGTC
CCTGGGGTCACTTTTCTGTCTGGAGGGCAGAGTGAGGAAGAGGCATCCATCAACCTCAACGCTATCAACAAGTGTCCCCT
GCTGAAGCCATGGGCCTTGACTTTCTCCTATGGCCGAGCCCTGCAGGCCTCTGCCCTAAAGGCCTGGGGTGGGAAAGAGG
AGAATCTGAAGGCAGCTCAGGAGGAGTACATCAAGAGAGCACTGGCCAACAGTCTCGCTTGTCAAGGAAAGTATACCCCA
AGTGGCAAGACTGGAGCCACAGCCAGCGAATCTCTCTTCATCTCTAACCATGCCTACTAA
ATGGCAACGCACAGGCAAGATGTGTCCATCTTCAACATGACCCGCCTGTCCCTGGCTATGGCTTTTTCCTTTCCCCCAGA
TGCCAATGAGCAACCCCACTCTGGGCTGGACAACACCCACCAACAGACAAAGGAGTTAGGAAAGGAAAGTACTACCACCG
GAACTATGCCCTGCCCATACCCAGCACTGACCACAGAGCAGAAGAAAGAGCTCTCTGACATCGCTCACCGAATTGTGGCT
CCAGGCAAGGGCATCCTGGCTGCAGATGAGTCCATTGGAAGCATGGGCAATCGCCTGCAGTCCATTGGCACTGAGAACAC
TGAAGAGAACAGGCGCTTCTTCCGCCAGTTGCTGCTGACTGCTGATGACCGTGTGAATCCCTGCATTGGGGGTGTGATCC
TCTTCCATGAGACACTGTACGAGAAGGCAGATGATGGACGTCCTTTCCCCCAAGTTATTAAGTCCAAGGGTGGTGTTGTG
GGCATTAAGGTAGATAAGGGTGTGGTGCCCCTGGCAGGAACCAATGGCGAGACTACTACCCAAGGGCTAGATGGGCTGTC
TGAACGCTGTGCCCAGTATAAGAAGGATGGAGCCGACTTTGCCAAGTGGCGCTGTGTGCTAAAGATTGGGAAACATACTC
CCTCGCCCCTTGCCATCATGGAGAATGCCAATGTTCTGGCCCGTTATGCCAGCATCTGCCAGCAGAATGGCATTGTACCC
ATTGTGGAGCCTGAAATTCTCCCTGATGGTGACCATGACTTGAGTTGTTGCCAGTATGTAACTGAGAAGGTACTGGCAGC
TGTCTACAAGGCTCTGAGTGACCACCATGTCTATCTGGAAGGCACATTGTTGAAGCCCAACATGGTCACCCCTGGTCATG
CTTGCACCCAGAAATTTTCCAATGAGGAGATTGCCATGGCAACAGTCACAGCACTTCGTCGCACAGTGCCACCTGCTGTC
CCTGGGGTCACTTTTCTGTCTGGAGGGCAGAGTGAGGAAGAGGCATCCATCAACCTCAACGCTATCAACAAGTGTCCCCT
GCTGAAGCCATGGGCCTTGACTTTCTCCTATGGCCGAGCCCTGCAGGCCTCTGCCCTAAAGGCCTGGGGTGGGAAAGAGG
AGAATCTGAAGGCAGCTCAGGAGGAGTACATCAAGAGAGCACTGGCCAACAGTCTCGCTTGTCAAGGAAAGTATACCCCA
AGTGGCAAGACTGGAGCCACAGCCAGCGAATCTCTCTTCATCTCTAACCATGCCTACTAA
ORF - retro_mmus_42 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.08 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MATRRPDGSSFNMTRLSLALAFSFPPVASEQPHSELGNTQQQT-ELGKESTATGTMPHPYPALTPEQKKE |
| MAT.R.D.S.FNMTRLSLA.AFSFPP.A.EQPHS.L.NT.QQT.ELGKEST.TGTMP.PYPALT.EQKKE | |
| Retrocopy | MATHRQDVSIFNMTRLSLAMAFSFPPDANEQPHSGLDNTHQQTKELGKESTTTGTMPCPYPALTTEQKKE |
| Parental | LSDIAHRIVAPGKGILAADESTGSIAKRLQSIGTENTEENRRFYRQLLLTADDRVNPCIGGVILFHETLY |
| LSDIAHRIVAPGKGILAADES.GS...RLQSIGTENTEENRRF.RQLLLTADDRVNPCIGGVILFHETLY | |
| Retrocopy | LSDIAHRIVAPGKGILAADESIGSMGNRLQSIGTENTEENRRFFRQLLLTADDRVNPCIGGVILFHETLY |
| Parental | QKADDGRPFPQVIKSKGGVVGIKVDKGVVPLAGTNGETTTQGLDGLSERCAQYKKDGADFAKWRCVLKIG |
| .KADDGRPFPQVIKSKGGVVGIKVDKGVVPLAGTNGETTTQGLDGLSERCAQYKKDGADFAKWRCVLKIG | |
| Retrocopy | EKADDGRPFPQVIKSKGGVVGIKVDKGVVPLAGTNGETTTQGLDGLSERCAQYKKDGADFAKWRCVLKIG |
| Parental | EHTPSALAIMENANVLARYASICQQNGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLE |
| .HTPS.LAIMENANVLARYASICQQNGIVPIVEPEILPDGDHDL..CQYVTEKVLAAVYKALSDHHVYLE | |
| Retrocopy | KHTPSPLAIMENANVLARYASICQQNGIVPIVEPEILPDGDHDLSCCQYVTEKVLAAVYKALSDHHVYLE |
| Parental | GTLLKPNMVTPGHACTQKFSNEEIAMATVTALRRTVPPAVTGVTFLSGGQSEEEASINLNAINKCPLLKP |
| GTLLKPNMVTPGHACTQKFSNEEIAMATVTALRRTVPPAV.GVTFLSGGQSEEEASINLNAINKCPLLKP | |
| Retrocopy | GTLLKPNMVTPGHACTQKFSNEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASINLNAINKCPLLKP |
| Parental | WALTFSYGRALQASALKAWGGKKENLKAAQEEYIKRALANSLACQGKYTPSGQSGAAASESLFISNHAY |
| WALTFSYGRALQASALKAWGGK.ENLKAAQEEYIKRALANSLACQGKYTPSG..GA.ASESLFISNHAY | |
| Retrocopy | WALTFSYGRALQASALKAWGGKEENLKAAQEEYIKRALANSLACQGKYTPSGKTGATASESLFISNHAY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .35 RPM | 1035 .35 RPM |
| SRP007412_cerebellum | 0 .22 RPM | 1045 .41 RPM |
| SRP007412_heart | 0 .97 RPM | 2044 .01 RPM |
| SRP007412_kidney | 0 .73 RPM | 1259 .68 RPM |
| SRP007412_liver | 0 .03 RPM | 87 .00 RPM |
| SRP007412_testis | 70 .30 RPM | 329 .87 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_42 was not detected
No EST(s) were mapped for retro_mmus_42 retrocopy.
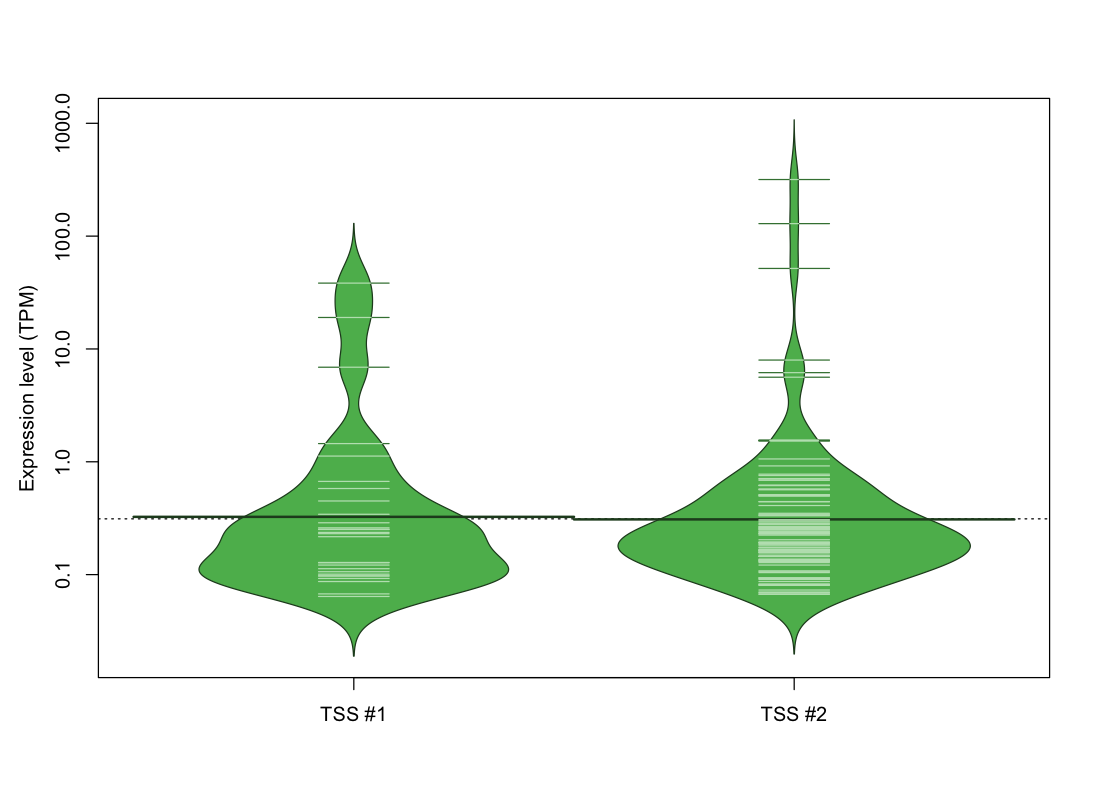
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_106377 | 1044 libraries | 23 libraries | 2 libraries | 1 library | 2 libraries |
| TSS #2 | TSS_106378 | 969 libraries | 92 libraries | 5 libraries | 3 libraries | 3 libraries |
The graphical summary, for retro_mmus_42 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_42 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_42 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 16 parental genes, and 30 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Canis familiaris | ENSCAFG00000008221 | 2 retrocopies | |
| Callithrix jacchus | ENSCJAG00000006874 | 1 retrocopy | |
| Cavia porcellus | ENSCPOG00000007434 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000019230 | 1 retrocopy | |
| Homo sapiens | ENSG00000149925 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000013979 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000011210 | 3 retrocopies | |
| Macaca mulatta | ENSMMUG00000000983 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000015696 | 4 retrocopies | |
| Mus musculus | ENSMUSG00000030695 | 3 retrocopies |
retro_mmus_2160, retro_mmus_42 , retro_mmus_90,
|
| Nomascus leucogenys | ENSNLEG00000002152 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000006329 | 1 retrocopy | |
| Ochotona princeps | ENSOPRG00000009760 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000007244 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000007983 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000023647 | 7 retrocopies |