RetrogeneDB ID: | retro_hsap_104 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 2:120979499..120980552(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000226479 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | TMEM185A | ||
| Ensembl ID: | ENSG00000155984 | ||
| Aliases: | TMEM185A, CXorf13, FAM11A, FRAXF, ee3 | ||
| Description: | transmembrane protein 185A [Source:HGNC Symbol;Acc:17125] |

Retrocopy-Parental alignment summary:
>retro_hsap_104
ATGAACCCCAGGGGCCTGTTCCAGGACTTCAACCCCAGTAAGTTTCTCATCTACACCTGCCTGCTGCTCTTCTCGGTGCT
GCTGCCCCTCCGCCTGGACGGCATCATCCAATGGAGCTACTGGGCCGTCTTTGCCCCCATATGGCTGTGGAAGCTTCTAG
TCGTCGCAGGCGCCTCCGTGGGCGCGGGCGTTTGGGCCCGCAACCCTCGCTACCGCACCGAGGGAGAGGCCTGTGTGGAG
TTCAAAGCCATGCTGATCGCTGTGGGCATCCACCTGCTGCTGCTCATGTTCGAAGTCCTGGTCTGCGACAGGGTGGAGAG
GGGCACCCACTTCTGGCTGCTGGTCTTCATGCCTCTCTTCTTCGTGTCCCCCGTGTCCGTGGCTGCCTGCGTCTGGGGCT
TTCGACACGATAGGTCGCTGGAGCTGGAGATCCTGTGCTCGGTCAACATCCTGCAGTTCATCTTCATCGCCCTAAAGCTG
GACAGGATTATTCACTGGCCGTGGCTGGTGGTGTTTGTGCCCCTGTGGATCCTCATGTCGTTCCTTTGCCTGGTCGTCCT
CTATTACATCGTCTGGTCCCTCCTGTTCCTGCGGTCCCTGGATGTGGTTGCCGAGCAGCGGAGAACACACGTGACCATGG
CTATCAGTTGGATAACGATTGTCGTGCCTCTGCTCACTTTTGAGGTCCTGCTGGTTCACAGATTGGATGGCCACAATACA
TTCTCCTACGTCTCCATATTTGTCCCCCTTTGGCTTTCCTTACTAACTTTAATGGCCACAACATTTAGGCGAAAGGGGGG
CAATCATTGGTGGTTTGGCATTCGCAGAGACTTCTGTCAGTTTCTGCTTGAAATTTTCCCATTTTTAAGAGAATATGGGA
ACATTTCATATGATCTCCATCACGAAGATAGTGAAGATGCTGAAGAAACATCAGTTCCAGAAGCTCCGAAAATTGCTCCA
ATATTTGGAAAGAAGGCCAGAGTAGTTATAACCCAGAGCCCTGGGAAATACGTTCCCCCCCCTCCCAAGTTAAATATTGA
TATGCCAGATTAA
ATGAACCCCAGGGGCCTGTTCCAGGACTTCAACCCCAGTAAGTTTCTCATCTACACCTGCCTGCTGCTCTTCTCGGTGCT
GCTGCCCCTCCGCCTGGACGGCATCATCCAATGGAGCTACTGGGCCGTCTTTGCCCCCATATGGCTGTGGAAGCTTCTAG
TCGTCGCAGGCGCCTCCGTGGGCGCGGGCGTTTGGGCCCGCAACCCTCGCTACCGCACCGAGGGAGAGGCCTGTGTGGAG
TTCAAAGCCATGCTGATCGCTGTGGGCATCCACCTGCTGCTGCTCATGTTCGAAGTCCTGGTCTGCGACAGGGTGGAGAG
GGGCACCCACTTCTGGCTGCTGGTCTTCATGCCTCTCTTCTTCGTGTCCCCCGTGTCCGTGGCTGCCTGCGTCTGGGGCT
TTCGACACGATAGGTCGCTGGAGCTGGAGATCCTGTGCTCGGTCAACATCCTGCAGTTCATCTTCATCGCCCTAAAGCTG
GACAGGATTATTCACTGGCCGTGGCTGGTGGTGTTTGTGCCCCTGTGGATCCTCATGTCGTTCCTTTGCCTGGTCGTCCT
CTATTACATCGTCTGGTCCCTCCTGTTCCTGCGGTCCCTGGATGTGGTTGCCGAGCAGCGGAGAACACACGTGACCATGG
CTATCAGTTGGATAACGATTGTCGTGCCTCTGCTCACTTTTGAGGTCCTGCTGGTTCACAGATTGGATGGCCACAATACA
TTCTCCTACGTCTCCATATTTGTCCCCCTTTGGCTTTCCTTACTAACTTTAATGGCCACAACATTTAGGCGAAAGGGGGG
CAATCATTGGTGGTTTGGCATTCGCAGAGACTTCTGTCAGTTTCTGCTTGAAATTTTCCCATTTTTAAGAGAATATGGGA
ACATTTCATATGATCTCCATCACGAAGATAGTGAAGATGCTGAAGAAACATCAGTTCCAGAAGCTCCGAAAATTGCTCCA
ATATTTGGAAAGAAGGCCAGAGTAGTTATAACCCAGAGCCCTGGGAAATACGTTCCCCCCCCTCCCAAGTTAAATATTGA
TATGCCAGATTAA
ORF - retro_hsap_104 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 88.57 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MNLRGLFQDFNPSKFLIYACLLLFSVLLALRLDGIIQWSYWAVFAPIWLWKLMVIVGASVGTGVWARNPQ |
| MN.RGLFQDFNPSKFLIY.CLLLFSVLL.LRLDGIIQWSYWAVFAPIWLWKL.V..GASVG.GVWARNP. | |
| Retrocopy | MNPRGLFQDFNPSKFLIYTCLLLFSVLLPLRLDGIIQWSYWAVFAPIWLWKLLVVAGASVGAGVWARNPR |
| Parental | YRAEGETCVEFKAMLIAVGIHLLLLMFEVLVCDRIERGSHFWLLVFMPLFFVSPVSVAACVWGFRHDRSL |
| YR.EGE.CVEFKAMLIAVGIHLLLLMFEVLVCDR.ERG.HFWLLVFMPLFFVSPVSVAACVWGFRHDRSL | |
| Retrocopy | YRTEGEACVEFKAMLIAVGIHLLLLMFEVLVCDRVERGTHFWLLVFMPLFFVSPVSVAACVWGFRHDRSL |
| Parental | ELEILCSVNILQFIFIALRLDKIIHWPWLVVCVPLWILMSFLCLVVLYYIVWSVLFLRSMDVIAEQRRTH |
| ELEILCSVNILQFIFIAL.LD.IIHWPWLVV.VPLWILMSFLCLVVLYYIVWS.LFLRS.DV.AEQRRTH | |
| Retrocopy | ELEILCSVNILQFIFIALKLDRIIHWPWLVVFVPLWILMSFLCLVVLYYIVWSLLFLRSLDVVAEQRRTH |
| Parental | ITMALSWMTIVVPLLTFEILLVHKLDGHNAFSSIPIFVPLWLSLITLMATTFGQKGGNHWWFGIRKDFCQ |
| .TMA.SW.TIVVPLLTFE.LLVH.LDGHN.FS...IFVPLWLSL.TLMATTF..KGGNHWWFGIR.DFCQ | |
| Retrocopy | VTMAISWITIVVPLLTFEVLLVHRLDGHNTFSYVSIFVPLWLSLLTLMATTFRRKGGNHWWFGIRRDFCQ |
| Parental | FLLEIFPFLREYGNISYDLHHEDNEETEETPVPEPPKIAPMFRKKARVVITQSPGKYVLPPPKLNIEMPD |
| FLLEIFPFLREYGNISYDLHHED.E..EET.VPE.PKIAP.F.KKARVVITQSPGKYV.PPPKLNI.MPD | |
| Retrocopy | FLLEIFPFLREYGNISYDLHHEDSEDAEETSVPEAPKIAPIFGKKARVVITQSPGKYVPPPPKLNIDMPD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 8 .98 RPM | 5 .59 RPM |
| bodymap2_adrenal | 11 .20 RPM | 5 .04 RPM |
| bodymap2_brain | 5 .93 RPM | 4 .81 RPM |
| bodymap2_breast | 9 .55 RPM | 4 .84 RPM |
| bodymap2_colon | 7 .08 RPM | 5 .39 RPM |
| bodymap2_heart | 8 .22 RPM | 7 .03 RPM |
| bodymap2_kidney | 9 .31 RPM | 5 .13 RPM |
| bodymap2_liver | 2 .66 RPM | 1 .31 RPM |
| bodymap2_lung | 7 .19 RPM | 2 .95 RPM |
| bodymap2_lymph_node | 7 .02 RPM | 5 .34 RPM |
| bodymap2_ovary | 12 .11 RPM | 10 .26 RPM |
| bodymap2_prostate | 9 .11 RPM | 6 .62 RPM |
| bodymap2_skeletal_muscle | 8 .77 RPM | 0 .91 RPM |
| bodymap2_testis | 14 .54 RPM | 9 .88 RPM |
| bodymap2_thyroid | 12 .69 RPM | 6 .89 RPM |
| bodymap2_white_blood_cells | 11 .75 RPM | 4 .98 RPM |
RNA Polymerase II actvity may be related with retro_hsap_104 in 46 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CFW | POLR2A | 2:120980443..120981326 |
| ENCFF002CFX | POLR2A | 2:120980458..120981320 |
| ENCFF002CGN | POLR2A | 2:120980622..120981068 |
| ENCFF002CHO | POLR2A | 2:120980378..120981566 |
| ENCFF002CIH | POLR2A | 2:120980851..120980999 |
| ENCFF002CIO | POLR2A | 2:120980595..120981052 |
| ENCFF002CJE | POLR2A | 2:120980703..120981074 |
| ENCFF002CJZ | POLR2A | 2:120980716..120981098 |
| ENCFF002CKX | POLR2A | 2:120980771..120981078 |
| ENCFF002CKX | POLR2A | 2:120981051..120981202 |
| ENCFF002CLM | POLR2A | 2:120980641..120981181 |
| ENCFF002CMI | POLR2A | 2:120980500..120981342 |
| ENCFF002COJ | POLR2A | 2:120980689..120981125 |
| ENCFF002CPG | POLR2A | 2:120980647..120981137 |
| ENCFF002CPH | POLR2A | 2:120980696..120981086 |
| ENCFF002CQA | POLR2A | 2:120980592..120981162 |
| ENCFF002CQC | POLR2A | 2:120980681..120981035 |
| ENCFF002CQC | POLR2A | 2:120980871..120981351 |
| ENCFF002CQE | POLR2A | 2:120980671..120981075 |
| ENCFF002CQE | POLR2A | 2:120980994..120981252 |
| ENCFF002CQG | POLR2A | 2:120980673..120981015 |
| ENCFF002CQI | POLR2A | 2:120980708..120981059 |
| ENCFF002CQK | POLR2A | 2:120980608..120981321 |
| ENCFF002CQM | POLR2A | 2:120980660..120981058 |
| ENCFF002CQO | POLR2A | 2:120980586..120981072 |
| ENCFF002CRK | POLR2A | 2:120980908..120981432 |
| ENCFF002CRO | POLR2A | 2:120980716..120981136 |
| ENCFF002CSY | POLR2A | 2:120980641..120981163 |
| ENCFF002CUP | POLR2A | 2:120980763..120981079 |
| ENCFF002CUQ | POLR2A | 2:120980676..120981180 |
| ENCFF002CVF | POLR2A | 2:120980709..120981125 |
| ENCFF002CVJ | POLR2A | 2:120980746..120981054 |
| ENCFF002CXM | POLR2A | 2:120980643..120981133 |
| ENCFF002CXN | POLR2A | 2:120980661..120981157 |
| ENCFF002CXO | POLR2A | 2:120980678..120981102 |
| ENCFF002CXP | POLR2A | 2:120980752..120981039 |
| ENCFF002CXR | POLR2A | 2:120980704..120981014 |
| ENCFF002CZC | POLR2A | 2:120980562..120981123 |
| ENCFF002CZD | POLR2A | 2:120980584..120981321 |
| ENCFF002DAE | POLR2A | 2:120980790..120981100 |
| ENCFF002DAE | POLR2A | 2:120981036..120981346 |
| ENCFF002DAH | POLR2A | 2:120980797..120981081 |
| ENCFF002DAK | POLR2A | 2:120980597..120981097 |
| ENCFF002DAV | POLR2A | 2:120980597..120981141 |
| ENCFF002DBB | POLR2A | 2:120980834..120981034 |
| ENCFF002DBE | POLR2A | 2:120980766..120981049 |
| ENCFF002DBO | POLR2A | 2:120980625..120981079 |
| ENCFF002DBP | POLR2A | 2:120980771..120981038 |
| ENCFF002DBQ | POLR2A | 2:120980743..120981113 |
| ENCFF002DBT | POLR2A | 2:120980748..120981118 |
5 EST(s) were mapped to retro_hsap_104 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA256930 | 120979906 | 120980225 | 100 | 319 | 0 | 319 |
| BE143874 | 120979779 | 120980045 | 99.7 | 265 | 1 | 264 |
| BF349607 | 120979557 | 120980049 | 95.8 | 470 | 3 | 454 |
| BF800502 | 120979772 | 120980109 | 100 | 336 | 0 | 335 |
| HY301666 | 120979535 | 120980044 | 100 | 509 | 0 | 509 |
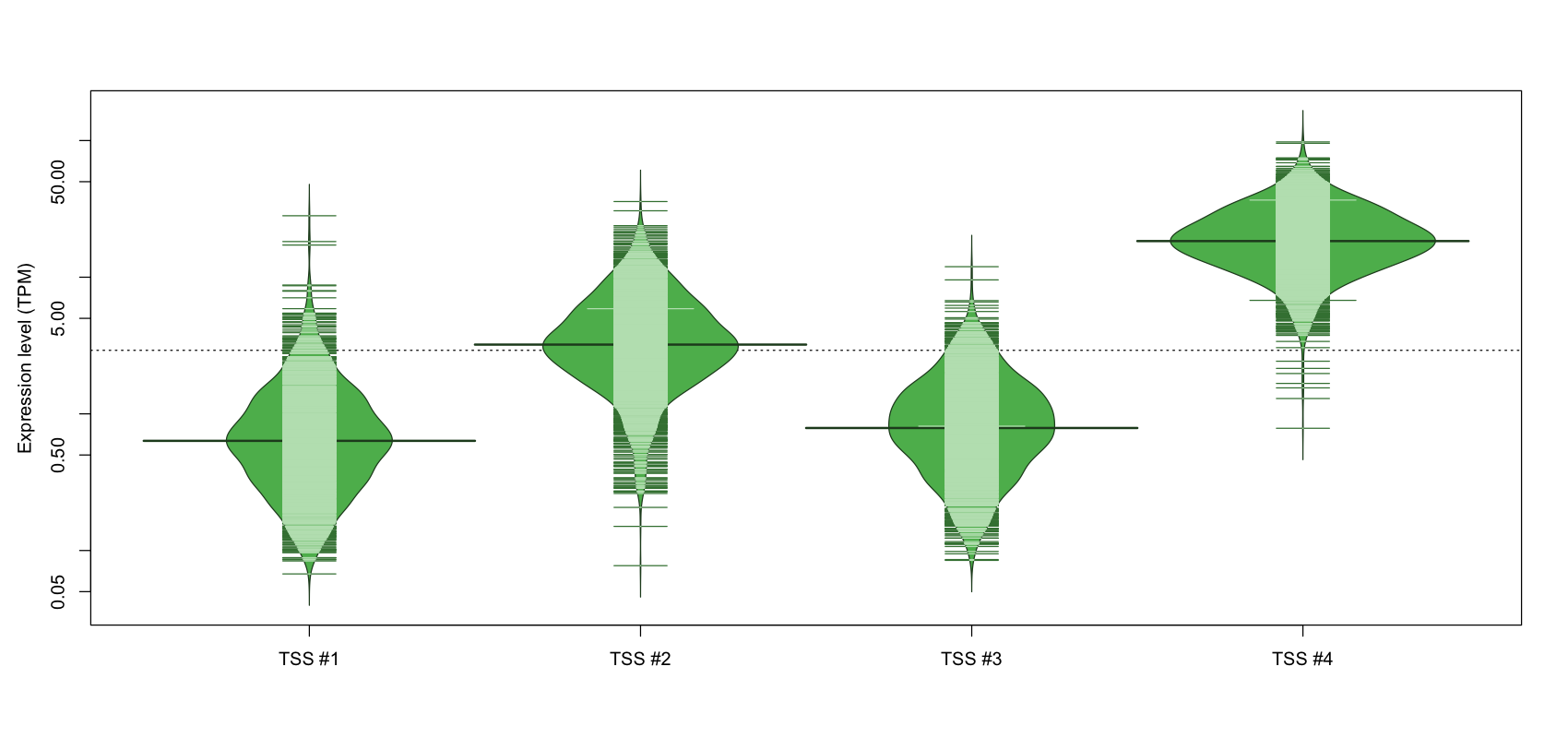
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_111735 | 686 libraries | 811 libraries | 315 libraries | 14 libraries | 3 libraries |
| TSS #2 | TSS_111736 | 114 libraries | 127 libraries | 1099 libraries | 367 libraries | 122 libraries |
| TSS #3 | TSS_111737 | 499 libraries | 795 libraries | 527 libraries | 7 libraries | 1 library |
| TSS #4 | TSS_111738 | 26 libraries | 1 library | 30 libraries | 173 libraries | 1599 libraries |
The graphical summary, for retro_hsap_104 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | TCGTGCCTCTGCTCACTTTT (20 nt long) |
| Reverse primer: | GCAAAGCCCAAGGAGAGAGT (20 nt long) |
| Anneling temperature: | 56 °C |
| (Expected) product size: | 883 |
| Electrophoresis gel image: | 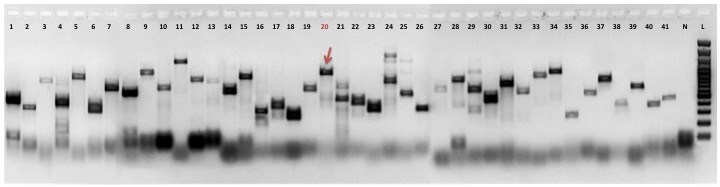 |
| Additional comment: | Red arrow indicates PCR product of retrogene in pooled cDNA from 16 human tissues (Clontech). Red number of lane indicates particular retrogene; L - GeneRuler 100 bp DNA Ladder (Thermo Fisher Scientific); N - No template control (water instead of cDNA) |
Retrocopy orthology:
Retrocopy retro_hsap_104 has 5 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_189 |
| Pongo abelii | retro_pabe_118 |
| Macaca mulatta | retro_mmul_310 |
| Mus musculus | retro_mmus_358 |
| Rattus norvegicus | retro_rnor_6 |
Parental genes homology:
Parental genes homology involve 26 parental genes, and 26 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 9 .67 RPM |
| CEU_NA11843 | 7 .54 RPM |
| CEU_NA11930 | 9 .39 RPM |
| CEU_NA12004 | 7 .03 RPM |
| CEU_NA12400 | 8 .17 RPM |
| CEU_NA12751 | 9 .81 RPM |
| CEU_NA12760 | 10 .73 RPM |
| CEU_NA12827 | 8 .44 RPM |
| CEU_NA12872 | 10 .51 RPM |
| CEU_NA12873 | 12 .41 RPM |
| FIN_HG00183 | 8 .37 RPM |
| FIN_HG00277 | 11 .20 RPM |
| FIN_HG00315 | 9 .03 RPM |
| FIN_HG00321 | 9 .28 RPM |
| FIN_HG00328 | 11 .22 RPM |
| FIN_HG00338 | 8 .41 RPM |
| FIN_HG00349 | 11 .13 RPM |
| FIN_HG00375 | 9 .17 RPM |
| FIN_HG00377 | 9 .43 RPM |
| FIN_HG00378 | 12 .83 RPM |
| GBR_HG00099 | 13 .25 RPM |
| GBR_HG00111 | 9 .61 RPM |
| GBR_HG00114 | 10 .76 RPM |
| GBR_HG00119 | 9 .32 RPM |
| GBR_HG00131 | 10 .26 RPM |
| GBR_HG00133 | 11 .05 RPM |
| GBR_HG00134 | 13 .34 RPM |
| GBR_HG00137 | 10 .64 RPM |
| GBR_HG00142 | 8 .70 RPM |
| GBR_HG00143 | 12 .16 RPM |
| TSI_NA20512 | 10 .48 RPM |
| TSI_NA20513 | 10 .69 RPM |
| TSI_NA20518 | 11 .80 RPM |
| TSI_NA20532 | 11 .22 RPM |
| TSI_NA20538 | 13 .64 RPM |
| TSI_NA20756 | 10 .68 RPM |
| TSI_NA20765 | 11 .04 RPM |
| TSI_NA20771 | 11 .06 RPM |
| TSI_NA20786 | 14 .83 RPM |
| TSI_NA20798 | 14 .41 RPM |
| YRI_NA18870 | 12 .49 RPM |
| YRI_NA18907 | 13 .21 RPM |
| YRI_NA18916 | 11 .85 RPM |
| YRI_NA19093 | 15 .23 RPM |
| YRI_NA19099 | 14 .14 RPM |
| YRI_NA19114 | 11 .90 RPM |
| YRI_NA19118 | 11 .64 RPM |
| YRI_NA19213 | 14 .52 RPM |
| YRI_NA19214 | 15 .25 RPM |
| YRI_NA19223 | 13 .65 RPM |