RetrogeneDB ID: | retro_hsap_57 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 9:6012289..6015607(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000137040 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | IPO5 | ||
| Ensembl ID: | ENSG00000065150 | ||
| Aliases: | IPO5, IMB3, KPNB3, Pse1, RANBP5, imp5 | ||
| Description: | importin 5 [Source:HGNC Symbol;Acc:6402] |

Retrocopy-Parental alignment summary:
>retro_hsap_57
ATGGCGGCAACCGCGTCTGCAGGGGTGCCGGCGACCGTGTCAGAAAAGCAAGAGTTTTACCAGCTTCTGAAGAACCTGAT
CAATCCAAGCTGTATGGTGCGGAGGCAAGCAGAGGAAATCTATGAAAATATCCCAGGTCTGTGTAAGACTACCTTCCTCT
TAGATGCCGTCAGAAATAGAAGAGCAGGTTATGAGGTGAGACAAATGGCTGCCGCACTGCTACGACGGCTTTTGTCCTCT
GGGTTTGAGGAGGTTTATCCAAATCTGCCTGCTGATGTTCAGAGAGATGTCAAGATTGAACTGATTCTGGCTGTTAAGTT
AGAAACACATGCTAGCATGAGGAAAAAACTTTGTGATATTTTTGCAGTGCTGGCCAGGAATTTGATAGATGAGGATGGCA
CTAACCACTGGCCGGAAGGTCTGAAGTTTCTTATTGATTCAATCTACTCCAAAAATGTGGTTCTATGGGAAGTTGCACTT
CACGTTTTCTGGCACTTTCCTGGGATTTTTGGGACCCAAGAGCGGCATGATTTGGATATCATCAAACGGTTGTTGGACCA
GTGTATTCAAGATCAAGAACATCCAGCAATCAGGACATTATCCGCTAGAGCTGCAGCTGCATTTGTACTTGCTAATGAGA
ATAATATTGCTCTTTTCAAAGACTTTGCAGACTTGCTTCCTGGAATCTTACAGGCTGTGAATGACTCATGCTACCAGGAT
GATGATTCAGTGCTAGAATCCCTTGTTGAGATTGCAGATACCGTACCTAAGTACTTGGGTCCTTATTTAGAAGATACTCT
ACAGTTGAGTTTGAAGTTATGTGGAGACTCTAGGCTTAGTAATCTGCAGCGCCAGCTGGCCCTCGAAGTTATAGTGACCT
TGTCTGAAACTGCCACTCCGATGTTGAAAAAACATACAAATATTATTGCACAGGCAGTTCCTCATATATTAGCAATGATG
GTTGATCTACAAGATGATGAGGACTGGGTAAATGCTGATGAAATGGAAGAAGATGATTTTGACAGCAATGCAGTTGCTGC
GGAGAGTGCACTAGACAGACTGGCTTGTGGGCTTGGTGGAAAAGTTGTTTTACCAATGACCAAGGAGCATATCATGCAGA
TGCTTCAGAGCCCTGACTGGAAGTATCGACATGCTGGATTAATGGCCTTATCTGCCATTGGAGAAGGATGCCATCAACAA
ATGGAATCAATTCTAGATGAAACAGTTAACTCCGTTTTGCTTTTTCTTCAGGATCCTCATCCAAGGGTGAGGGCTGCAGC
CTGTACTACACTTGGACAGATGGCTACAGATTTTGCACCTAATTTCCAAAAGAAATTTCATGAAACAGTGATTGCAGCTC
TGTTACGTACCATGGAAAATCAAGGTAATCAGCGTGTGCAATCACATGCAGCTTCTGCTCTTATTATTTTTATTGAAGAC
TGCCCTAAATCATTGCTAGTTCTATATGTGGATAGTATGGTGAAAAATCTACATTCCGTCTTGGTGATTAAACTTCAAGA
GTTGATTCGGAATGGAACTAAGTTGGCTTTGGAACAACTTGTGACAACCATTGCATCAGTTGCAGATACAATAGAAGAAA
AATTTGTCCCATATTATGATATATTCATGCCCTCACTAAAGCACATTGTTGAGCTTGCTGTTCAGAAGGAACTCAAGCTT
CTGAGAGGAAAAACTATCGAGTGCATTAGCCATATTGGTCTTGCTGTTGGGAAGGAAAAATTTATGCAAGATGCATCAAA
TGTGATGCAGCTGTTGTTGAAGACACAATCAGACTTAAATAATATGGAAGATGATGACCCTCAGACCTCTTACATGGTTT
CAGCATGGGCTAGAATGTGTAAAATTCTTGGAAAAGATTTTCAACAGTACCTTCCACTGGTTATCGAGCCTCTTATTAAG
ACTGCTTCAGCTAAACCTGATGTTGCTCTCTTAGACACACAGGATGTGGAAAATATGAGTGACGATGATGGCTGGCAATT
TGTAAATCTTGGAGACCAGCAGAGTTTTGGAATTAAAACTTCAGGACTTGAAGCAAAAGCAACTGCTTGCCAAATGTTGG
TTTACTATGCTAAGGAGTTAAGGGAAGGGTTTGTGGAATATACAGAACAAGTTGTGAAGCTGATGGTTCCTTTACTGAAA
TTTTATTTCCATGACAATGTTCGAGTGGCAGCAGCAGAGTCCATGCCTTTTCTCCTGGAATGTGCAAGAATTCGTGGCCC
AGAGTATCTTGCACAGATGTGGCAATTCATATGTGACCCCTTAATCAAGGCTATTGGTACTGAACCAGATACAGATGTGC
TCTCAGAAATAATGAATTCTTTTGCAAAGTCCATTGAAGTTATGGGAGATGGTTGCCTTAATGATGAACACTTGGAAGAA
CTGGGAGGAATACTGAAAGCAAAACTTGAAGGGCACTTTAAAAACCAAGAATTGAGACAGGTGAAAAGACAGGAAGAAAA
CTATGATCAACAGGTTGAGATGTCTCTGCAAGATGAGGATGAATGTGATGTTTATATTCTGACCAAAGTATCAGATATTT
TGCACTCATTATTTAGTACTTATAAGGAAAAGATTTTACCATGGTTTGAACAACTACTTCCATTAATTGTAAATCTAATT
TGTTCAAGTAGGCCATGGCCAGACAGACAGTGGGGATTGTGCATATTTGATGACATCATAGAGCACTGCAGTCCAACTTC
ATTTAAATATGTAGAATATTTTCGGTGGCCAATGCTACTAAATATGCGAGATAACAACCCTGAAGTCAGGCAAGCTGCTG
CTTATGGCCTGGGTGTCATGGCACAGTTTGGTGGAGATGATTATCGTTCTTTATGTTCAGAAGCTGTTCCACTTCTGGTA
AAAGTTATTAAGTGTGCAAATTCCAAAACCAAAAAAAATGTCATTGCTACAGAGAACTGTATCTCAGCAATAGGGAAGAT
TTTGAAGTTTAAGCCTAACTGTGTAAATGTAGATGAAGTTCTTCCACACTGGTTATCATGGCTTCCACTGCATGAAGATA
AAGAGGAAGCTATTCAGACTTTGAGTTTTCTCTGTGACCTAATTGAAAGTAACCACCCAGTTGTAATTGGTCCAAATAAT
TCCAATCTTCCCAAAATAATCAGTATAATTGCAGAAGGAAAAATTAATGAGACTATTAACTATGAGGATCCTTGTGCCAA
ACGCCTAGCTAATGTCGTGCGTCAGGTACAGACTTCTGAAGATTTATGGTTGGAATGTGTATCACAACTTGATGATGAAC
AGCAGGAAGCCTTACAGGAGTTGCTAAATTTTGCTTGA
ATGGCGGCAACCGCGTCTGCAGGGGTGCCGGCGACCGTGTCAGAAAAGCAAGAGTTTTACCAGCTTCTGAAGAACCTGAT
CAATCCAAGCTGTATGGTGCGGAGGCAAGCAGAGGAAATCTATGAAAATATCCCAGGTCTGTGTAAGACTACCTTCCTCT
TAGATGCCGTCAGAAATAGAAGAGCAGGTTATGAGGTGAGACAAATGGCTGCCGCACTGCTACGACGGCTTTTGTCCTCT
GGGTTTGAGGAGGTTTATCCAAATCTGCCTGCTGATGTTCAGAGAGATGTCAAGATTGAACTGATTCTGGCTGTTAAGTT
AGAAACACATGCTAGCATGAGGAAAAAACTTTGTGATATTTTTGCAGTGCTGGCCAGGAATTTGATAGATGAGGATGGCA
CTAACCACTGGCCGGAAGGTCTGAAGTTTCTTATTGATTCAATCTACTCCAAAAATGTGGTTCTATGGGAAGTTGCACTT
CACGTTTTCTGGCACTTTCCTGGGATTTTTGGGACCCAAGAGCGGCATGATTTGGATATCATCAAACGGTTGTTGGACCA
GTGTATTCAAGATCAAGAACATCCAGCAATCAGGACATTATCCGCTAGAGCTGCAGCTGCATTTGTACTTGCTAATGAGA
ATAATATTGCTCTTTTCAAAGACTTTGCAGACTTGCTTCCTGGAATCTTACAGGCTGTGAATGACTCATGCTACCAGGAT
GATGATTCAGTGCTAGAATCCCTTGTTGAGATTGCAGATACCGTACCTAAGTACTTGGGTCCTTATTTAGAAGATACTCT
ACAGTTGAGTTTGAAGTTATGTGGAGACTCTAGGCTTAGTAATCTGCAGCGCCAGCTGGCCCTCGAAGTTATAGTGACCT
TGTCTGAAACTGCCACTCCGATGTTGAAAAAACATACAAATATTATTGCACAGGCAGTTCCTCATATATTAGCAATGATG
GTTGATCTACAAGATGATGAGGACTGGGTAAATGCTGATGAAATGGAAGAAGATGATTTTGACAGCAATGCAGTTGCTGC
GGAGAGTGCACTAGACAGACTGGCTTGTGGGCTTGGTGGAAAAGTTGTTTTACCAATGACCAAGGAGCATATCATGCAGA
TGCTTCAGAGCCCTGACTGGAAGTATCGACATGCTGGATTAATGGCCTTATCTGCCATTGGAGAAGGATGCCATCAACAA
ATGGAATCAATTCTAGATGAAACAGTTAACTCCGTTTTGCTTTTTCTTCAGGATCCTCATCCAAGGGTGAGGGCTGCAGC
CTGTACTACACTTGGACAGATGGCTACAGATTTTGCACCTAATTTCCAAAAGAAATTTCATGAAACAGTGATTGCAGCTC
TGTTACGTACCATGGAAAATCAAGGTAATCAGCGTGTGCAATCACATGCAGCTTCTGCTCTTATTATTTTTATTGAAGAC
TGCCCTAAATCATTGCTAGTTCTATATGTGGATAGTATGGTGAAAAATCTACATTCCGTCTTGGTGATTAAACTTCAAGA
GTTGATTCGGAATGGAACTAAGTTGGCTTTGGAACAACTTGTGACAACCATTGCATCAGTTGCAGATACAATAGAAGAAA
AATTTGTCCCATATTATGATATATTCATGCCCTCACTAAAGCACATTGTTGAGCTTGCTGTTCAGAAGGAACTCAAGCTT
CTGAGAGGAAAAACTATCGAGTGCATTAGCCATATTGGTCTTGCTGTTGGGAAGGAAAAATTTATGCAAGATGCATCAAA
TGTGATGCAGCTGTTGTTGAAGACACAATCAGACTTAAATAATATGGAAGATGATGACCCTCAGACCTCTTACATGGTTT
CAGCATGGGCTAGAATGTGTAAAATTCTTGGAAAAGATTTTCAACAGTACCTTCCACTGGTTATCGAGCCTCTTATTAAG
ACTGCTTCAGCTAAACCTGATGTTGCTCTCTTAGACACACAGGATGTGGAAAATATGAGTGACGATGATGGCTGGCAATT
TGTAAATCTTGGAGACCAGCAGAGTTTTGGAATTAAAACTTCAGGACTTGAAGCAAAAGCAACTGCTTGCCAAATGTTGG
TTTACTATGCTAAGGAGTTAAGGGAAGGGTTTGTGGAATATACAGAACAAGTTGTGAAGCTGATGGTTCCTTTACTGAAA
TTTTATTTCCATGACAATGTTCGAGTGGCAGCAGCAGAGTCCATGCCTTTTCTCCTGGAATGTGCAAGAATTCGTGGCCC
AGAGTATCTTGCACAGATGTGGCAATTCATATGTGACCCCTTAATCAAGGCTATTGGTACTGAACCAGATACAGATGTGC
TCTCAGAAATAATGAATTCTTTTGCAAAGTCCATTGAAGTTATGGGAGATGGTTGCCTTAATGATGAACACTTGGAAGAA
CTGGGAGGAATACTGAAAGCAAAACTTGAAGGGCACTTTAAAAACCAAGAATTGAGACAGGTGAAAAGACAGGAAGAAAA
CTATGATCAACAGGTTGAGATGTCTCTGCAAGATGAGGATGAATGTGATGTTTATATTCTGACCAAAGTATCAGATATTT
TGCACTCATTATTTAGTACTTATAAGGAAAAGATTTTACCATGGTTTGAACAACTACTTCCATTAATTGTAAATCTAATT
TGTTCAAGTAGGCCATGGCCAGACAGACAGTGGGGATTGTGCATATTTGATGACATCATAGAGCACTGCAGTCCAACTTC
ATTTAAATATGTAGAATATTTTCGGTGGCCAATGCTACTAAATATGCGAGATAACAACCCTGAAGTCAGGCAAGCTGCTG
CTTATGGCCTGGGTGTCATGGCACAGTTTGGTGGAGATGATTATCGTTCTTTATGTTCAGAAGCTGTTCCACTTCTGGTA
AAAGTTATTAAGTGTGCAAATTCCAAAACCAAAAAAAATGTCATTGCTACAGAGAACTGTATCTCAGCAATAGGGAAGAT
TTTGAAGTTTAAGCCTAACTGTGTAAATGTAGATGAAGTTCTTCCACACTGGTTATCATGGCTTCCACTGCATGAAGATA
AAGAGGAAGCTATTCAGACTTTGAGTTTTCTCTGTGACCTAATTGAAAGTAACCACCCAGTTGTAATTGGTCCAAATAAT
TCCAATCTTCCCAAAATAATCAGTATAATTGCAGAAGGAAAAATTAATGAGACTATTAACTATGAGGATCCTTGTGCCAA
ACGCCTAGCTAATGTCGTGCGTCAGGTACAGACTTCTGAAGATTTATGGTTGGAATGTGTATCACAACTTGATGATGAAC
AGCAGGAAGCCTTACAGGAGTTGCTAAATTTTGCTTGA
ORF - retro_hsap_57 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 79.07 % |
| Parental protein coverage: | 98.57 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | SAMAAAAAEQQQFYLLLGNLLSPDNVVRKQAEETYENIPGQSKITFLLQAIRNTTAAEEARQMAAVLLRR |
| ....A...E.Q.FY.LL.NL..P...VR.QAEE.YENIPG..K.TFLL.A.RN..A..E.RQMAA.LLRR | |
| Retrocopy | AGVPATVSEKQEFYQLLKNLINPSCMVRRQAEEIYENIPGLCKTTFLLDAVRNRRAGYEVRQMAAALLRR |
| Parental | LLSSAFDEVYPALPSDVQTAIKSELLMIIQMETQSSMRKKVCDIAAELARNLIDEDGNNQWPEGLKFLFD |
| LLSS.F.EVYP.LP.DVQ...K.EL......ET..SMRKK.CDI.A.LARNLIDEDG.N.WPEGLKFL.D | |
| Retrocopy | LLSSGFEEVYPNLPADVQRDVKIELILAVKLETHASMRKKLCDIFAVLARNLIDEDGTNHWPEGLKFLID |
| Parental | SVSSQNVGLREAALHIFWNFPGIFGNQQQHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNV |
| S..S.NV.L.E.ALH.FW.FPGIFG.Q..H.LD.IKR.L.QC.QDQEHP.IRTLSARA.AAF.LANE.N. | |
| Retrocopy | SIYSKNVVLWEVALHVFWHFPGIFGTQERHDLDIIKRLLDQCIQDQEHPAIRTLSARAAAAFVLANENNI |
| Parental | ALFKHFADLLPGFLQAVNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTSLNNMQRQL |
| ALFK.FADLLPG.LQAVNDSCYQ.DDSVL.SLVEIADTVPKYL.P.LE.TLQLSLKLCGD..L.N.QRQL | |
| Retrocopy | ALFKDFADLLPGILQAVNDSCYQDDDSVLESLVEIADTVPKYLGPYLEDTLQLSLKLCGDSRLSNLQRQL |
| Parental | ALEVIVTLSETAAAMLRKHTNIVAQTIPQMLAMMVDLEEDEDWANADELEDDDFDSNAVAGESALDRMAC |
| ALEVIVTLSETA..ML.KHTNI.AQ..P..LAMMVDL..DEDW.NADE.E.DDFDSNAVA.ESALDR.AC | |
| Retrocopy | ALEVIVTLSETATPMLKKHTNIIAQAVPHILAMMVDLQDDEDWVNADEMEEDDFDSNAVAAESALDRLAC |
| Parental | GLGGKLVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCHQQMEGILNEIVNFVLLFLQDPHPRVRYA |
| GLGGK.VLPM.KEHIMQMLQ.PDWKYRHAGLMALSAIGEGCHQQME.IL.E.VN.VLLFLQDPHPRVR.A | |
| Retrocopy | GLGGKVVLPMTKEHIMQMLQSPDWKYRHAGLMALSAIGEGCHQQMESILDETVNSVLLFLQDPHPRVRAA |
| Parental | ACNAVGQMATDFAPGFQKKFHEKVIAALLQTMEDQGNQRVQAHAAAALINFTEDCPKSLLIPYLDNLVKH |
| AC...GQMATDFAP.FQKKFHE.VIAALL.TME.QGNQRVQ.HAA.ALI.F.EDCPKSLL..Y.D..VK. | |
| Retrocopy | ACTTLGQMATDFAPNFQKKFHETVIAALLRTMENQGNQRVQSHAASALIIFIEDCPKSLLVLYVDSMVKN |
| Parental | LHSIMVLKLQELIQKGTKLVLEQVVTSIASVADTAEEKFVPYYDLFMPSLKHIVENAVQKELRLLRGKTI |
| LHS..V.KLQELI..GTKL.LEQ.VT.IASVADT.EEKFVPYYD.FMPSLKHIVE.AVQKEL.LLRGKTI | |
| Retrocopy | LHSVLVIKLQELIRNGTKLALEQLVTTIASVADTIEEKFVPYYDIFMPSLKHIVELAVQKELKLLRGKTI |
| Parental | ECISLIGLAVGKEKFMQDASDVMQLLLKTQTDFNDMEDDDPQISYMISAWARMCKILGKEFQQYLPVVMG |
| ECIS.IGLAVGKEKFMQDAS.VMQLLLKTQ.D.N.MEDDDPQ.SYM.SAWARMCKILGK.FQQYLP.V.. | |
| Retrocopy | ECISHIGLAVGKEKFMQDASNVMQLLLKTQSDLNNMEDDDPQTSYMVSAWARMCKILGKDFQQYLPLVIE |
| Parental | PLMKTASIKPEVALLDTQDMENMSDDDGWEFVNLGDQQSFGIKTAGLEEKSTACQMLVCYAKELKEGFVE |
| PL.KTAS.KP.VALLDTQD.ENMSDDDGW.FVNLGDQQSFGIKT.GLE.K.TACQMLV.YAKEL.EGFVE | |
| Retrocopy | PLIKTASAKPDVALLDTQDVENMSDDDGWQFVNLGDQQSFGIKTSGLEAKATACQMLVYYAKELREGFVE |
| Parental | YTEQVVKLMVPLLKFYFHDGVRVAAAESMPLLLECARVRGPEYLTQMWHFMCDALIKAIGTEPDSDVLSE |
| YTEQVVKLMVPLLKFYFHD.VRVAAAESMP.LLECAR.RGPEYL.QMW.F.CD.LIKAIGTEPD.DVLSE | |
| Retrocopy | YTEQVVKLMVPLLKFYFHDNVRVAAAESMPFLLECARIRGPEYLAQMWQFICDPLIKAIGTEPDTDVLSE |
| Parental | IMHSFAKCIEVMGDGCLNNEHFEELGGILKAKLEEHFKNQELRQVKRQDEDYDEQVEESLQDEDDNDVYI |
| IM.SFAK.IEVMGDGCLN.EH.EELGGILKAKLE.HFKNQELRQVKRQ.E.YD.QVE.SLQDED..DVYI | |
| Retrocopy | IMNSFAKSIEVMGDGCLNDEHLEELGGILKAKLEGHFKNQELRQVKRQEENYDQQVEMSLQDEDECDVYI |
| Parental | LTKVSDILHSIFSSYKEKVLPWFEQLLPLIVNLICPHRPWPDRQWGLCIFDDVIEHCSPASFKYAEYFLR |
| LTKVSDILHS.FS.YKEK.LPWFEQLLPLIVNLIC..RPWPDRQWGLCIFDD.IEHCSP.SFKY.EYF.. | |
| Retrocopy | LTKVSDILHSLFSTYKEKILPWFEQLLPLIVNLICSSRPWPDRQWGLCIFDDIIEHCSPTSFKYVEYFRW |
| Parental | PMLQYVCDNSPEVRQAAAYGLGVMAQYGGDNYRPFCTEALPLLVRVIQSADSKTKENVNATENCISAVGK |
| PML....DN.PEVRQAAAYGLGVMAQ.GGD.YR..C.EA.PLLV.VI..A.SKTK.NV.ATENCISA.GK | |
| Retrocopy | PMLLNMRDNNPEVRQAAAYGLGVMAQFGGDDYRSLCSEAVPLLVKVIKCANSKTKKNVIATENCISAIGK |
| Parental | IMKFKPDCVNVEEVLPHWLSWLPLHEDKEEAVQTFNYLCDLIESNHPIVLGPNNTNLPKIFSIIAEGEMH |
| I.KFKP.CVNV.EVLPHWLSWLPLHEDKEEA.QT...LCDLIESNHP.V.GPNN.NLPKI.SIIAEG... | |
| Retrocopy | ILKFKPNCVNVDEVLPHWLSWLPLHEDKEEAIQTLSFLCDLIESNHPVVIGPNNSNLPKIISIIAEGKIN |
| Parental | EAIKHEDPCAKRLANVVRQVQTSGGLWTECIAQLSPEQQAAIQELLNSA |
| E.I..EDPCAKRLANVVRQVQTS..LW.EC..QL..EQQ.A.QELLN.A | |
| Retrocopy | ETINYEDPCAKRLANVVRQVQTSEDLWLECVSQLDDEQQEALQELLNFA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 12 .38 RPM | 222 .71 RPM |
| bodymap2_adrenal | 25 .63 RPM | 178 .80 RPM |
| bodymap2_brain | 45 .19 RPM | 200 .23 RPM |
| bodymap2_breast | 20 .06 RPM | 219 .09 RPM |
| bodymap2_colon | 11 .72 RPM | 250 .15 RPM |
| bodymap2_heart | 24 .61 RPM | 162 .49 RPM |
| bodymap2_kidney | 27 .20 RPM | 147 .64 RPM |
| bodymap2_liver | 9 .12 RPM | 63 .33 RPM |
| bodymap2_lung | 7 .63 RPM | 171 .10 RPM |
| bodymap2_lymph_node | 30 .72 RPM | 164 .28 RPM |
| bodymap2_ovary | 30 .11 RPM | 371 .79 RPM |
| bodymap2_prostate | 13 .67 RPM | 267 .56 RPM |
| bodymap2_skeletal_muscle | 6 .98 RPM | 426 .43 RPM |
| bodymap2_testis | 17 .51 RPM | 602 .34 RPM |
| bodymap2_thyroid | 39 .41 RPM | 260 .98 RPM |
| bodymap2_white_blood_cells | 35 .52 RPM | 132 .36 RPM |
RNA Polymerase II actvity may be related with retro_hsap_57 in 49 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CFW | POLR2A | 9:6015438..6015882 |
| ENCFF002CFX | POLR2A | 9:6015454..6015896 |
| ENCFF002CGN | POLR2A | 9:6015494..6015872 |
| ENCFF002CHO | POLR2A | 9:6015388..6015950 |
| ENCFF002CIH | POLR2A | 9:6015338..6015842 |
| ENCFF002CIO | POLR2A | 9:6015457..6015894 |
| ENCFF002CJE | POLR2A | 9:6015336..6015965 |
| ENCFF002CJZ | POLR2A | 9:6015408..6015901 |
| ENCFF002CKX | POLR2A | 9:6015467..6015901 |
| ENCFF002CLM | POLR2A | 9:6015494..6016034 |
| ENCFF002CMI | POLR2A | 9:6015399..6015928 |
| ENCFF002COJ | POLR2A | 9:6015369..6015805 |
| ENCFF002CPG | POLR2A | 9:6015677..6015863 |
| ENCFF002CPH | POLR2A | 9:6015424..6015814 |
| ENCFF002CQA | POLR2A | 9:6015652..6015932 |
| ENCFF002CQC | POLR2A | 9:6015489..6015868 |
| ENCFF002CQE | POLR2A | 9:6015441..6015882 |
| ENCFF002CQG | POLR2A | 9:6015509..6015776 |
| ENCFF002CQI | POLR2A | 9:6015576..6015836 |
| ENCFF002CQK | POLR2A | 9:6015443..6015901 |
| ENCFF002CQM | POLR2A | 9:6015476..6015867 |
| ENCFF002CQO | POLR2A | 9:6015489..6015876 |
| ENCFF002CRK | POLR2A | 9:6015482..6016006 |
| ENCFF002CSY | POLR2A | 9:6015380..6015816 |
| ENCFF002CUP | POLR2A | 9:6015551..6015673 |
| ENCFF002CUQ | POLR2A | 9:6015602..6015860 |
| ENCFF002CVJ | POLR2A | 9:6015278..6015888 |
| ENCFF002CXM | POLR2A | 9:6015503..6015993 |
| ENCFF002CXN | POLR2A | 9:6015354..6015850 |
| ENCFF002CXO | POLR2A | 9:6015700..6015858 |
| ENCFF002CXP | POLR2A | 9:6015492..6015875 |
| ENCFF002CXP | POLR2A | 9:6015492..6015880 |
| ENCFF002CXR | POLR2A | 9:6015493..6015871 |
| ENCFF002CZC | POLR2A | 9:6015395..6015757 |
| ENCFF002CZC | POLR2A | 9:6015671..6015909 |
| ENCFF002CZD | POLR2A | 9:6015417..6015716 |
| ENCFF002CZD | POLR2A | 9:6015553..6016063 |
| ENCFF002CZQ | POLR2A | 9:6015422..6015905 |
| ENCFF002CZW | POLR2A | 9:6015394..6015803 |
| ENCFF002CZW | POLR2A | 9:6015394..6015819 |
| ENCFF002CZY | POLR2A | 9:6015422..6015937 |
| ENCFF002DAE | POLR2A | 9:6015524..6015834 |
| ENCFF002DAH | POLR2A | 9:6015499..6015783 |
| ENCFF002DAK | POLR2A | 9:6015496..6015996 |
| ENCFF002DAS | POLR2A | 9:6015384..6015860 |
| ENCFF002DAV | POLR2A | 9:6015332..6015876 |
| ENCFF002DAY | POLR2A | 9:6015467..6016103 |
| ENCFF002DBB | POLR2A | 9:6015618..6015818 |
| ENCFF002DBB | POLR2A | 9:6014872..6015072 |
| ENCFF002DBE | POLR2A | 9:6015487..6015860 |
| ENCFF002DBO | POLR2A | 9:6015527..6015699 |
| ENCFF002DBP | POLR2A | 9:6015566..6015663 |
| ENCFF002DBQ | POLR2A | 9:6015554..6015924 |
| ENCFF002DBT | POLR2A | 9:6015409..6015779 |
45 EST(s) were mapped to retro_hsap_57 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA515855 | 6012496 | 6012904 | 99.8 | 407 | 1 | 406 |
| AI187237 | 6012238 | 6012691 | 100 | 453 | 0 | 453 |
| AL527784 | 6012299 | 6013279 | 98.6 | 883 | 12 | 867 |
| AU137263 | 6012177 | 6012971 | 98.9 | 781 | 7 | 770 |
| AW403296 | 6012505 | 6012838 | 100 | 333 | 0 | 333 |
| BE004761 | 6013601 | 6014090 | 99.2 | 488 | 1 | 486 |
| BE311758 | 6012745 | 6013559 | 99.2 | 799 | 4 | 782 |
| BE708609 | 6012712 | 6013008 | 99.7 | 293 | 1 | 290 |
| BG180616 | 6012830 | 6013521 | 98.7 | 680 | 3 | 663 |
| BG260670 | 6012354 | 6012991 | 99.3 | 635 | 2 | 632 |
| BM145818 | 6012689 | 6013148 | 98.7 | 445 | 4 | 436 |
| BM456277 | 6012250 | 6012970 | 98 | 714 | 4 | 702 |
| BM839500 | 6013450 | 6013801 | 100 | 351 | 0 | 351 |
| BQ067780 | 6013826 | 6014490 | 98.1 | 658 | 6 | 649 |
| BQ953326 | 6012234 | 6012977 | 99.1 | 735 | 1 | 731 |
| BX327088 | 6012747 | 6013617 | 96.3 | 812 | 14 | 781 |
| BX391578 | 6012923 | 6013770 | 97.2 | 803 | 16 | 770 |
| BX436545 | 6012345 | 6013115 | 99.1 | 745 | 4 | 740 |
| BX471142 | 6013029 | 6013451 | 99.8 | 421 | 1 | 420 |
| DA010013 | 6012723 | 6013266 | 99.9 | 542 | 1 | 541 |
| DA100319 | 6013593 | 6014140 | 100 | 547 | 0 | 547 |
| DA114707 | 6014054 | 6014646 | 100 | 592 | 0 | 592 |
| DA116213 | 6013590 | 6014180 | 100 | 590 | 0 | 590 |
| DA125312 | 6013938 | 6014502 | 99.5 | 564 | 0 | 563 |
| DA155272 | 6013262 | 6013831 | 100 | 569 | 0 | 569 |
| DA195769 | 6013618 | 6014176 | 100 | 558 | 0 | 558 |
| DA224982 | 6014357 | 6014923 | 100 | 566 | 0 | 566 |
| DA248958 | 6014331 | 6014912 | 99.7 | 578 | 2 | 575 |
| DA297861 | 6012692 | 6013264 | 99.2 | 572 | 0 | 570 |
| DA394083 | 6014279 | 6014866 | 99.9 | 586 | 1 | 585 |
| DA401152 | 6013812 | 6014372 | 100 | 560 | 0 | 560 |
| DA500733 | 6013083 | 6013657 | 99.2 | 574 | 0 | 572 |
| DA516052 | 6013472 | 6014024 | 99.1 | 552 | 0 | 550 |
| DA547886 | 6012825 | 6013072 | 100 | 247 | 0 | 247 |
| DA603626 | 6012379 | 6012843 | 100 | 464 | 0 | 464 |
| DA805153 | 6013857 | 6014522 | 100 | 665 | 0 | 665 |
| DA806690 | 6014328 | 6014889 | 100 | 561 | 0 | 561 |
| DB137963 | 6013425 | 6013962 | 100 | 537 | 0 | 537 |
| DB152826 | 6013662 | 6014224 | 100 | 562 | 0 | 562 |
| DB233454 | 6014090 | 6014678 | 100 | 588 | 0 | 588 |
| DB288409 | 6014399 | 6014951 | 100 | 552 | 0 | 552 |
| DC420670 | 6012592 | 6013157 | 99.5 | 565 | 0 | 564 |
| DC426897 | 6014018 | 6014510 | 98.4 | 480 | 4 | 472 |
| H05851 | 6012281 | 6012670 | 95.6 | 380 | 0 | 369 |
| N87727 | 6014499 | 6014783 | 98.3 | 277 | 4 | 269 |
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_193522 | 58 libraries | 14 libraries | 203 libraries | 478 libraries | 1076 libraries |
The graphical summary, for retro_hsap_57 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
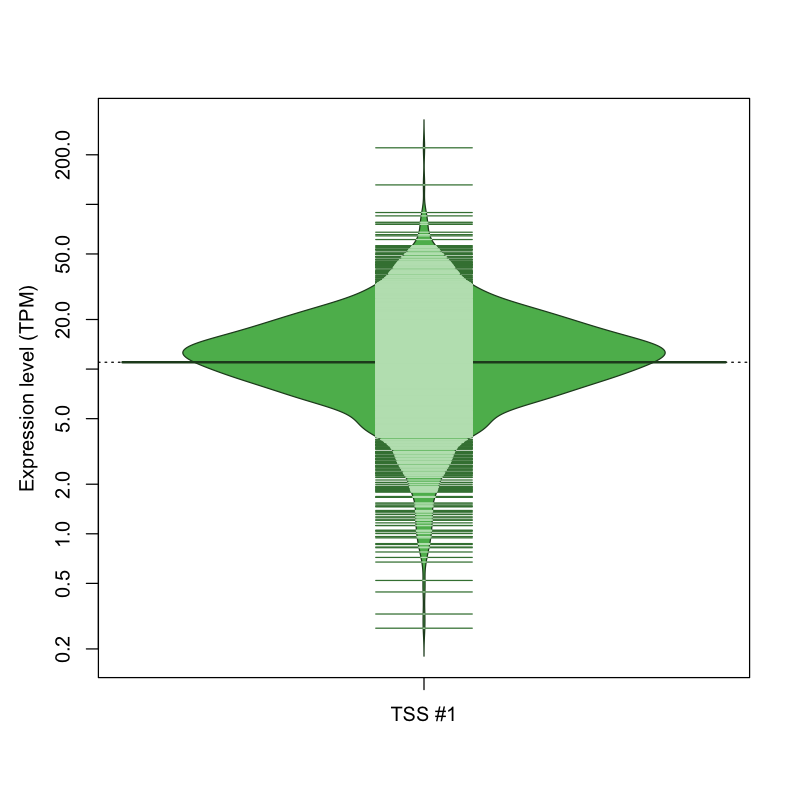
retro_hsap_57 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_57 has 4 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_49 |
| Gorilla gorilla | retro_ggor_47 |
| Mus musculus | retro_mmus_251 |
| Rattus norvegicus | retro_rnor_240 |
Parental genes homology:
Parental genes homology involve 21 parental genes, and 25 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 21 .96 RPM |
| CEU_NA11843 | 13 .93 RPM |
| CEU_NA11930 | 28 .52 RPM |
| CEU_NA12004 | 18 .80 RPM |
| CEU_NA12400 | 18 .10 RPM |
| CEU_NA12751 | 32 .05 RPM |
| CEU_NA12760 | 19 .36 RPM |
| CEU_NA12827 | 22 .92 RPM |
| CEU_NA12872 | 26 .89 RPM |
| CEU_NA12873 | 26 .74 RPM |
| FIN_HG00183 | 29 .54 RPM |
| FIN_HG00277 | 25 .62 RPM |
| FIN_HG00315 | 24 .29 RPM |
| FIN_HG00321 | 28 .59 RPM |
| FIN_HG00328 | 24 .60 RPM |
| FIN_HG00338 | 22 .42 RPM |
| FIN_HG00349 | 21 .94 RPM |
| FIN_HG00375 | 27 .56 RPM |
| FIN_HG00377 | 19 .79 RPM |
| FIN_HG00378 | 30 .05 RPM |
| GBR_HG00099 | 30 .28 RPM |
| GBR_HG00111 | 18 .23 RPM |
| GBR_HG00114 | 26 .43 RPM |
| GBR_HG00119 | 30 .10 RPM |
| GBR_HG00131 | 33 .31 RPM |
| GBR_HG00133 | 23 .03 RPM |
| GBR_HG00134 | 27 .41 RPM |
| GBR_HG00137 | 16 .88 RPM |
| GBR_HG00142 | 28 .76 RPM |
| GBR_HG00143 | 28 .18 RPM |
| TSI_NA20512 | 12 .72 RPM |
| TSI_NA20513 | 35 .35 RPM |
| TSI_NA20518 | 27 .23 RPM |
| TSI_NA20532 | 29 .08 RPM |
| TSI_NA20538 | 32 .39 RPM |
| TSI_NA20756 | 20 .19 RPM |
| TSI_NA20765 | 32 .41 RPM |
| TSI_NA20771 | 27 .03 RPM |
| TSI_NA20786 | 19 .01 RPM |
| TSI_NA20798 | 24 .11 RPM |
| YRI_NA18870 | 22 .75 RPM |
| YRI_NA18907 | 21 .94 RPM |
| YRI_NA18916 | 23 .81 RPM |
| YRI_NA19093 | 11 .64 RPM |
| YRI_NA19099 | 20 .82 RPM |
| YRI_NA19114 | 23 .15 RPM |
| YRI_NA19118 | 20 .12 RPM |
| YRI_NA19213 | 29 .20 RPM |
| YRI_NA19214 | 26 .86 RPM |
| YRI_NA19223 | 29 .47 RPM |