RetrogeneDB ID: | retro_mmus_251 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 19:29809632..29812950(-) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000074909 | |
| Aliases: | Ranbp6, C630001B19 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Ipo5 | ||
| Ensembl ID: | ENSMUSG00000030662 | ||
| Aliases: | Ipo5, 1110011C18Rik, 5730478E03Rik, AA409333, C76941, IMB3, Kpnb3, Ranbp5 | ||
| Description: | importin 5 [Source:MGI Symbol;Acc:MGI:1917822] |

Retrocopy-Parental alignment summary:
>retro_mmus_251
ATGGCGGCGGCCGGGTCTGCAGGGTTGCCGGCGACCGTGTCGGAAAAGCAGGAGTTTTACCAGCTTCTGAAGAACCTGAT
CAATCCAAGCTGCATGGTGCGGAGGCAAGCCGAGGAGGTCTATGAGAATATCCCAGGTCTGTGTAAGACTACATTTCTCT
TAGACGCCGTCCGAAATAGAAGAGCAGGTTATGAGGTGAGACAAATGGCTGCCGCACTGCTACGACGGCTTTTGTCCTCA
GGGTTTGAGGAGGTCTATCCAAATCTACCTCCTGAAGTGCAGAGGGATGTCAAGATTGAACTGATACTGGCCGTTAAGTT
AGAAACACATGCTAGCATGAGGAAAAAGCTTTGTGATATTTTTGCGGTGCTGGCCAGGAATTTGATAGATGAGAGTGGCA
CTAACCATTGGCCAGAAGGTCTGAAATTCCTCATTGATTCGATTCACTCTAAAAATGTGGTTCTGTGGGAAGTTGCACTT
CACGTTTTCTGGCACTTTCCTGGGATTTTTGGGAACCAGGATCGTCACGATTTGGATATCATCAAGAGGTTGTTGGACCA
GTGTATTCAAGATCAGGAGCATCCAGCAATCAGGACATTATCTGCTAGAGCTGCAGCTACGTTCGTCCTTGCTAATGAAA
ACAATATTGCTCTTTTCAAAGACTTTGCGGATTTGCTTCCTGGGATCTTACAGGCTGTGAATGATTCCTGCTACCAGGAT
GATGATTCTGTGCTAGAATCCCTTGTTGAGATTGCAGATACTGTACCTAAGTACTTGGGTCCCTATCTAGAAGATACTCT
GCAGTTGAGCCTGAAGTTGTGTGGAGACTCTAGGCTTAGTAACCTGCAACGCCAGCTGGCTCTGGAAGTAATAGTCACCT
TATCTGAAACTGCAACTCCAATGTTGAAAAAGCATACGAATATTATTGCACAGGCTGTGCCTCATATATTAGCAATGATG
GTTGATCTTCAAGATGATGATGACTGGGTAAATGCTGATGAAATGGAAGAAGATGACTTTGACAGCAATGCGGTTGCTGC
TGAGAGTGCCCTAGACAGACTGGCTTGTGGACTTGGTGGGAAAGTTGTTTTGCCAATGACCAAGGAGCATATCATGCAGA
TGCTTCAGAGCCCTGACTGGAAATGTCGACACGCCGGATTAATGGCCTTATCAGCCATTGGAGAAGGTTGCCATCAGCAA
ATGGAACCAATTCTAGATGAAACAGTTAACTCGGTTTTACTTTTTCTTCAGGATCCTCATCCAAGGGTAAGGGCTGCTGC
CTGTACTACACTTGGACAGATGGCTACAGATTTTGCACCTAGTTTCCAAAAGAAATTCCATGAGATAGTGATTACAGCTT
TGTTGCGAACCATGGAAAACCAAGGTAATCAACGTGTGCAATCTCACGCAGCTTCTGCTCTTGTTATTTTTATTGAAGAC
TGCCCCAAGTCATTGCTAATTCTATATTTGGAAAATATGGTAAAAAGTCTCCATTCCATCTTGGTAATTAAACTTCAAGA
GCTGATTCGGAATGGAACTAAGTTGGCCTTAGAACAACTTGTGACAACCATTGCTTCCGTTGCAGATGCAATAGAAGAAA
GCTTTATTCCATATTATGATATATTTATGCCCTCACTCAAACACGTTGTTGAGCTCGCTGTTCAAAAGGAACTCAAGCTT
CTTCGAGGAAAAACTATTGAATGCATTAGCCATGTTGGACTTGCCGTTGGGAAGGAAAAATTTATGCAAGATGCATCAAA
TGTGATGCAGTTACTGTTGAAAACACAATCAGACTTAAATAACATGGAAGATGATGACCCTCAGACTTCTTACATGGTTT
CAGCCTGGGCTAGAATGTGTAAAATCCTTGGCAAAGACTTTGAGCAATATCTCCCACTGGTTATTGAGCCACTTATCAAG
ACTGCTTCAGCTAAGCCTGATGTCGCTCTCTTAGACACACAGGATGTGGAGAACATGAGTGACGACGATGGCTGGCAGTT
TGTAAACCTCGGAGACCAGCAAAGCTTTGGCATCAAAACTTCAGGACTTGAAGCAAAAGCAACTGCTTGCCAGATGTTGG
TTTACTATGCGAAGGAGTTAAGAGAAGGATTTGTGGAATACACGGAACAAGTTGTGAAGATGATGGTTCCTCTGCTGAAA
TTTTATTTTCATGACAATGTTCGAGTGGCAGCTGCGGAAGCCATGCCCTTTCTCTTGGAATGTGCAAGAATTCGTGGCTC
AGAGTATCTTTCACAGATGTGGCAGTTTATATGTGATCCCTTAATCAAGGCAATTGGAACTGAACCTGATACAGATGTAC
TCTCAGAAATAATGAATTCTTTTGCCAAGTCCATTGAGGTCATGGGAGATGGTTGCCTCAACGATGAACACTTGGAAGAA
CTGGGAGGAATTTTGAAGGCAAAACTTGAAGGGCACTTTAAAAACCAAGAGTTACGGCAAGTAAAAAGGCAAGAAGAAAA
TTATGACCAACAGGTTGAAATGTCTCTTCAAGATGAGGATGAATGTGATGTTTATATTCTGACCAAAGTATCAGATATTT
TGCACTCATTATTTAGTACTTACAAGGAAAAAATTTTGCCATGGTTTGAGCAGCTACTCCCACTAATTGTAAATCTAATT
TGTTCAAGTAGGCCATGGCCAGACAGACAATGGGGACTATGCATATTTGATGACATCATAGAGCATTGCAGTCCAACCTC
ATTCAAGTATGTTGAGTATTTCCGGTGGCCGATGCTTCTCAATATGCGAGACAACAACCCTGAAGTCAGACAAGCTGCTG
CTTATGGACTGGGAGTCATGGCACAGTTTGGTGGAGATGACTACCGTTCCTTGTGTTCAGAAGCCGTTCCACTCTTGGTA
AAAGTTATTAAGTGTGCAAATTCCAAAACCAAAAAGAATGTGATTGCTACAGAGAACTGTATCTCAGCAATCGGAAAGAT
TCTGAAGTTTAAGCCCAACTGTGTAAATGTGGATGAAGTTCTCCCACACTGGTTATCGTGGCTTCCGCTGCATGAAGATA
AAGAAGAAGCTATTCAGACTTTGAATTTTCTCTGTGACCTAATTGAAAGCAACCACCCAGTTGTGATTGGTCCAAATAAT
TCCAATCTTCCCAAAATCATCAGTATAATCGCAGAAGGAAAAATTAATGAAACTATTAGCCATGAAGACCCCTGTGCCAA
ACGCCTAGCTAATGTTGTCCGTCAGATACAGACTTCTGAAGAATTGTGGTTGGAATGCACATCTCAACTCGATGATGAGC
AGCAGGAAGCTTTACATGAGTTACTCAGTTTTGCTTGA
ORF - retro_mmus_251 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 77.58 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAAAAAEQQQFYLLLGNLLSPDNVVRKQAEETYENIPGRSKITFLLQAIRNTTAAEEARQMAAVLLRRLL |
| ..A...E.Q.FY.LL.NL..P...VR.QAEE.YENIPG..K.TFLL.A.RN..A..E.RQMAA.LLRRLL | |
| Retrocopy | LPATVSEKQEFYQLLKNLINPSCMVRRQAEEVYENIPGLCKTTFLLDAVRNRRAGYEVRQMAAALLRRLL |
| Parental | SSAFDEVYPALPSDVQTAIKSELLMIIQMETQSSMRKKICDIAAELARNLIDEDGNNQWPEGLKFLFDSV |
| SS.F.EVYP.LP..VQ...K.EL......ET..SMRKK.CDI.A.LARNLIDE.G.N.WPEGLKFL.DS. | |
| Retrocopy | SSGFEEVYPNLPPEVQRDVKIELILAVKLETHASMRKKLCDIFAVLARNLIDESGTNHWPEGLKFLIDSI |
| Parental | SSQNMGLREAALHIFWNFPGIFGNQQQHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNVAL |
| .S.N..L.E.ALH.FW.FPGIFGNQ..H.LD.IKR.L.QC.QDQEHP.IRTLSARA.A.F.LANE.N.AL | |
| Retrocopy | HSKNVVLWEVALHVFWHFPGIFGNQDRHDLDIIKRLLDQCIQDQEHPAIRTLSARAAATFVLANENNIAL |
| Parental | FKHFADLLPGFLQAVNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTNLNNMQRQLAL |
| FK.FADLLPG.LQAVNDSCYQ.DDSVL.SLVEIADTVPKYL.P.LE.TLQLSLKLCGD..L.N.QRQLAL | |
| Retrocopy | FKDFADLLPGILQAVNDSCYQDDDSVLESLVEIADTVPKYLGPYLEDTLQLSLKLCGDSRLSNLQRQLAL |
| Parental | EVIVTLSETAAAMLRKHTSLIAQTIPQMLAMMVDLEEDEDWANADELEDDDFDSNAVAGESALDRMACGL |
| EVIVTLSETA..ML.KHT..IAQ..P..LAMMVDL..D.DW.NADE.E.DDFDSNAVA.ESALDR.ACGL | |
| Retrocopy | EVIVTLSETATPMLKKHTNIIAQAVPHILAMMVDLQDDDDWVNADEMEEDDFDSNAVAAESALDRLACGL |
| Parental | GGKLVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCHQQMEGILNEIVNFVLLFLQDPHPRVRYAAC |
| GGK.VLPM.KEHIMQMLQ.PDWK.RHAGLMALSAIGEGCHQQME.IL.E.VN.VLLFLQDPHPRVR.AAC | |
| Retrocopy | GGKVVLPMTKEHIMQMLQSPDWKCRHAGLMALSAIGEGCHQQMEPILDETVNSVLLFLQDPHPRVRAAAC |
| Parental | NAVGQMATDFAPGFQKKFHEKVIAALLQTMEDQGNQRVQAHAAAALINFTEDCPKSLLIPYLDNLVKHLH |
| ...GQMATDFAP.FQKKFHE.VI.ALL.TME.QGNQRVQ.HAA.AL..F.EDCPKSLLI.YL.N.VK.LH | |
| Retrocopy | TTLGQMATDFAPSFQKKFHEIVITALLRTMENQGNQRVQSHAASALVIFIEDCPKSLLILYLENMVKSLH |
| Parental | SIMVLKLQELIQKGTKLVLEQVVTSIASVADTAEEKFVPYYDLFMPSLKHIVENAVQKELRLLRGKTIEC |
| SI.V.KLQELI..GTKL.LEQ.VT.IASVAD..EE.F.PYYD.FMPSLKH.VE.AVQKEL.LLRGKTIEC | |
| Retrocopy | SILVIKLQELIRNGTKLALEQLVTTIASVADAIEESFIPYYDIFMPSLKHVVELAVQKELKLLRGKTIEC |
| Parental | ISLIGLAVGKEKFMQDASDVMQLLLKTQTDFNDMEDDDPQISYMISAWARMCKILGKEFQQYLPVVMGPL |
| IS..GLAVGKEKFMQDAS.VMQLLLKTQ.D.N.MEDDDPQ.SYM.SAWARMCKILGK.F.QYLP.V..PL | |
| Retrocopy | ISHVGLAVGKEKFMQDASNVMQLLLKTQSDLNNMEDDDPQTSYMVSAWARMCKILGKDFEQYLPLVIEPL |
| Parental | MKTASIKPEVALLDTQDMENMSDDDGWEFVNLGDQQSFGIKTAGLEEKSTACQMLVCYAKELKEGFVEYT |
| .KTAS.KP.VALLDTQD.ENMSDDDGW.FVNLGDQQSFGIKT.GLE.K.TACQMLV.YAKEL.EGFVEYT | |
| Retrocopy | IKTASAKPDVALLDTQDVENMSDDDGWQFVNLGDQQSFGIKTSGLEAKATACQMLVYYAKELREGFVEYT |
| Parental | EQVVKLMVPLLKFYFHDGVRVAAAESMPLLLECARVRGPEYLTQMWHFMCDALIKAIGTEPDSDVLSEIM |
| EQVVK.MVPLLKFYFHD.VRVAAAE.MP.LLECAR.RG.EYL.QMW.F.CD.LIKAIGTEPD.DVLSEIM | |
| Retrocopy | EQVVKMMVPLLKFYFHDNVRVAAAEAMPFLLECARIRGSEYLSQMWQFICDPLIKAIGTEPDTDVLSEIM |
| Parental | HSFAKCIEVMGDGCLNNEHFEELGGILKAKLEEHFKNQELRQVKRQDEDYDEQVEESLQDEDDNDVYILT |
| .SFAK.IEVMGDGCLN.EH.EELGGILKAKLE.HFKNQELRQVKRQ.E.YD.QVE.SLQDED..DVYILT | |
| Retrocopy | NSFAKSIEVMGDGCLNDEHLEELGGILKAKLEGHFKNQELRQVKRQEENYDQQVEMSLQDEDECDVYILT |
| Parental | KVSDILHSIFSSYKEKVLPWFEQLLPLIVNLICPQRPWPDRQWGLCIFDDIVEHCSPASFKYAEYFISPM |
| KVSDILHS.FS.YKEK.LPWFEQLLPLIVNLIC..RPWPDRQWGLCIFDDI.EHCSP.SFKY.EYF..PM | |
| Retrocopy | KVSDILHSLFSTYKEKILPWFEQLLPLIVNLICSSRPWPDRQWGLCIFDDIIEHCSPTSFKYVEYFRWPM |
| Parental | LQYVCDNSPEVRQAAAYGLGVMAQFGGDNYRPFCTDALPLLVRVIQAPEAKTKENVNATENCISAVGKIM |
| L....DN.PEVRQAAAYGLGVMAQFGGD.YR..C..A.PLLV.VI.....KTK.NV.ATENCISA.GKI. | |
| Retrocopy | LLNMRDNNPEVRQAAAYGLGVMAQFGGDDYRSLCSEAVPLLVKVIKCANSKTKKNVIATENCISAIGKIL |
| Parental | KFKPDCVNVEEVLPHWLSWLPLHEDKEEAVQTFSYLCDLIESNHPIVLGPNNTNLPKIFSIIAEGEMHEA |
| KFKP.CVNV.EVLPHWLSWLPLHEDKEEA.QT...LCDLIESNHP.V.GPNN.NLPKI.SIIAEG...E. | |
| Retrocopy | KFKPNCVNVDEVLPHWLSWLPLHEDKEEAIQTLNFLCDLIESNHPVVIGPNNSNLPKIISIIAEGKINET |
| Parental | IKHEDPCAKRLANVVRQVQTSGGLWTECIAQLSPEQQAAIQELLNSA |
| I.HEDPCAKRLANVVRQ.QTS..LW.EC..QL..EQQ.A..ELL..A | |
| Retrocopy | ISHEDPCAKRLANVVRQIQTSEELWLECTSQLDDEQQEALHELLSFA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 35 .16 RPM | 164 .99 RPM |
| SRP007412_cerebellum | 27 .66 RPM | 119 .06 RPM |
| SRP007412_heart | 12 .23 RPM | 209 .91 RPM |
| SRP007412_kidney | 14 .48 RPM | 124 .58 RPM |
| SRP007412_liver | 10 .08 RPM | 73 .05 RPM |
| SRP007412_testis | 39 .02 RPM | 1253 .84 RPM |
RNA Polymerase II activity may be related with retro_mmus_251 in 3 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 19:29812725..29813239 |
| ENCFF001YIJ | POLR2A | 19:29812712..29813322 |
| ENCFF001YKA | POLR2A | 19:29812509..29813425 |
No EST(s) were mapped for retro_mmus_251 retrocopy.
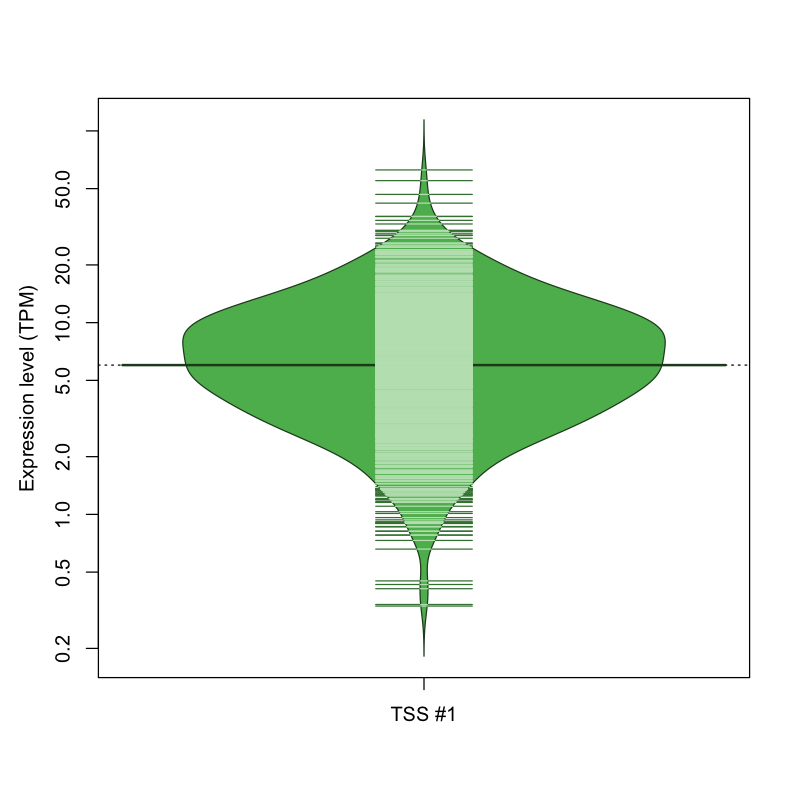
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_63613 | 42 libraries | 19 libraries | 381 libraries | 356 libraries | 274 libraries |
The graphical summary, for retro_mmus_251 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_251 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_251 has 4 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Homo sapiens | retro_hsap_57 |
| Pan troglodytes | retro_ptro_49 |
| Gorilla gorilla | retro_ggor_47 |
| Rattus norvegicus | retro_rnor_240 |