RetrogeneDB ID: | retro_mmus_3356 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 9:75374608..75375811(-) | ||
| Located in intron of: | ENSMUSG00000042688 | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tuba3b | ||
| Ensembl ID: | ENSMUSG00000067338 | ||
| Aliases: | Tuba3b, M[a]7, Tuba7 | ||
| Description: | tubulin, alpha 3B [Source:MGI Symbol;Acc:MGI:1095408] |

Retrocopy-Parental alignment summary:
>retro_mmus_3356
GGAGACGACTCATTCAACACATTCTTCAGTGAGACTGCAGCTGGCACGCATGTGCCCCAGCAGTGTTTGTGGACCTGGAG
CCAACTGTGGTTGATGAAGTGCACACTGGAACCTACTGGCAGCTTTTCCACCCAAAGCAGCGGATCACTGGGAAGGAAGA
TGCTGGCAACAATTAAGCCAGAGGTCGTTATACAATTGGCAAGAAGATTGTTGACCTGGTCCTGGACAGGATCTGAAAAC
TGGCCGATCTGTGCACGGGACTTCAGGGCTTCCTCATCTTCCACAGCTTTGGAGGTAGCACAGGGTCTGTGTTTGCATCT
CTGCTGATGAAGCAGCTCTCAGCGGATTATGGCAAGAAGTCCAAGCTGGAGTTTGCCATTTACCCAGCCCCACAGGTTTC
TACAGCTGTCGTGGAGCCTTACAACTCCATCCTGACCACGCACACAACACTGGAACATTCCGACTGTGCTTTCATGGTGG
ATAATGAAGCCATCTACGACATCTGCCTGTGCAGCCTGGATATTGAACGTCCCACATACACCAACCTCAATCGTCTGATT
GGGCAGATTGTGTCATCCATCACAGCCCCCCTGAGGTTTGATGGCGCCCCGAATGTGGACTTAACAGAATTCCAGACTAA
CCTGGTGCCATATCCTCACATCCACTTCCCACTGGCAACCTATGCCCCAGTCATCTCTGCTGAGAAGGCATACCATGAGC
AGAGATCAGTGGCAGAGATCACCAATGCTTGCTTCGAGCCAGCCAATCAGATGGTCAAGTGTGACTTTCGTCACAGCAAA
TACACGGCCTGCTGCATGTTGTACAGGGGGATGTGGTTCCCAAAGATGTCAATGCACTGTTGCCACCATCAAGACCAAGC
TTGTAGATTGGTGCTGGACTGGATTTAAGGTGGGTATTAACTATCAGCCTTCTACTGTGGTCCCTGGGGGAGACCTGGCC
AAGGTGCAGCGGGCCGTGTGCATGCTGAGCAATACCACTGCCATTGCAGAGTCCTGGGCCCACCTGGACCACAAATTTGA
CCTCATGTACGCCAAGCGGGCCTTTGTGCATTGGTACGTGGGAGAAGGCATGGAGGAAGGGGAGTTCTCCGAGGCCCGGG
AGGACCTGGCAGCGCTGGAGAAGGATTATGAAGAGGTGGGCGTGGATTCCGTGGGAGCAGAGGCAGAAGAAGGGGAGGAG
TAC
GGAGACGACTCATTCAACACATTCTTCAGTGAGACTGCAGCTGGCACGCATGTGCCCCAGCAGTGTTTGTGGACCTGGAG
CCAACTGTGGTTGATGAAGTGCACACTGGAACCTACTGGCAGCTTTTCCACCCAAAGCAGCGGATCACTGGGAAGGAAGA
TGCTGGCAACAATTAAGCCAGAGGTCGTTATACAATTGGCAAGAAGATTGTTGACCTGGTCCTGGACAGGATCTGAAAAC
TGGCCGATCTGTGCACGGGACTTCAGGGCTTCCTCATCTTCCACAGCTTTGGAGGTAGCACAGGGTCTGTGTTTGCATCT
CTGCTGATGAAGCAGCTCTCAGCGGATTATGGCAAGAAGTCCAAGCTGGAGTTTGCCATTTACCCAGCCCCACAGGTTTC
TACAGCTGTCGTGGAGCCTTACAACTCCATCCTGACCACGCACACAACACTGGAACATTCCGACTGTGCTTTCATGGTGG
ATAATGAAGCCATCTACGACATCTGCCTGTGCAGCCTGGATATTGAACGTCCCACATACACCAACCTCAATCGTCTGATT
GGGCAGATTGTGTCATCCATCACAGCCCCCCTGAGGTTTGATGGCGCCCCGAATGTGGACTTAACAGAATTCCAGACTAA
CCTGGTGCCATATCCTCACATCCACTTCCCACTGGCAACCTATGCCCCAGTCATCTCTGCTGAGAAGGCATACCATGAGC
AGAGATCAGTGGCAGAGATCACCAATGCTTGCTTCGAGCCAGCCAATCAGATGGTCAAGTGTGACTTTCGTCACAGCAAA
TACACGGCCTGCTGCATGTTGTACAGGGGGATGTGGTTCCCAAAGATGTCAATGCACTGTTGCCACCATCAAGACCAAGC
TTGTAGATTGGTGCTGGACTGGATTTAAGGTGGGTATTAACTATCAGCCTTCTACTGTGGTCCCTGGGGGAGACCTGGCC
AAGGTGCAGCGGGCCGTGTGCATGCTGAGCAATACCACTGCCATTGCAGAGTCCTGGGCCCACCTGGACCACAAATTTGA
CCTCATGTACGCCAAGCGGGCCTTTGTGCATTGGTACGTGGGAGAAGGCATGGAGGAAGGGGAGTTCTCCGAGGCCCGGG
AGGACCTGGCAGCGCTGGAGAAGGATTATGAAGAGGTGGGCGTGGATTCCGTGGGAGCAGAGGCAGAAGAAGGGGAGGAG
TAC
ORF - retro_mmus_3356 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 89.73 % |
| Parental protein coverage: | 90.22 % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected | 3 |
Retrocopy - Parental Gene Alignment:
| Parental | GDDSFNTFFSETGAGKHVPR-AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKE |
| GDDSFNTFFSET.AG.HVP..AVFVDLEPTVVDEV.TGTY.QLFHP.Q.ITGKEDA.NN.ARG.YTIGK. | |
| Retrocopy | GDDSFNTFFSETAAGTHVPQ<AVFVDLEPTVVDEVHTGTYWQLFHPKQRITGKEDAGNN*ARGRYTIGKK |
| Parental | IVDLVLDRIRKLADLCTGLQGFLIFHSFGGGTGSGFASLLMERLSVDYGKKSKLEFAIYPAPQVSTAVVE |
| IVDLVLDRI.KLADLCTGLQGFLIFHSFGG.TGS.FASLLM..LS.DYGKKSKLEFAIYPAPQVSTAVVE | |
| Retrocopy | IVDLVLDRI*KLADLCTGLQGFLIFHSFGGSTGSVFASLLMKQLSADYGKKSKLEFAIYPAPQVSTAVVE |
| Parental | PYNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRFDGALNVDLT |
| PYNSILTTHTTLEHSDCAFMVDNEAIYDIC...LDIERPTYTNLNRLIGQIVSSITA.LRFDGA.NVDLT | |
| Retrocopy | PYNSILTTHTTLEHSDCAFMVDNEAIYDICLCSLDIERPTYTNLNRLIGQIVSSITAPLRFDGAPNVDLT |
| Parental | EFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQMVKCDPRHGKYMACCMLYR-GD |
| EFQTNLVPYP.IHFPLATYAPVISAEKAYHEQ.SVAEITNACFEPANQMVKCD.RH.KY.ACCMLYR.GD | |
| Retrocopy | EFQTNLVPYPHIHFPLATYAPVISAEKAYHEQRSVAEITNACFEPANQMVKCDFRHSKYTACCMLYR<GD |
| Parental | VVPKDVNA-AIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVPGGDLAKVQRAVCMLSNTTAIAEAWARL |
| VVPKDVNA...ATIKTK.....VDWC.TGFKVGINYQP.TVVPGGDLAKVQRAVCMLSNTTAIAE.WA.L | |
| Retrocopy | VVPKDVNA<TVATIKTK----LVDWCWTGFKVGINYQPSTVVPGGDLAKVQRAVCMLSNTTAIAESWAHL |
| Parental | DHKFDLMYAKRAFVHWYVGEGMEEGEFSEAREDLAALEKDYEEVGVDSVEAEAEEGEEY |
| DHKFDLMYAKRAFVHWYVGEGMEEGEFSEAREDLAALEKDYEEVGVDSV.AEAEEGEEY | |
| Retrocopy | DHKFDLMYAKRAFVHWYVGEGMEEGEFSEAREDLAALEKDYEEVGVDSVGAEAEEGEEY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 0 .07 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 0 .00 RPM |
| SRP007412_heart | 0 .00 RPM | 0 .09 RPM |
| SRP007412_kidney | 0 .00 RPM | 0 .15 RPM |
| SRP007412_liver | 0 .00 RPM | 0 .11 RPM |
| SRP007412_testis | 0 .69 RPM | 924 .43 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_3356 was not detected
No EST(s) were mapped for retro_mmus_3356 retrocopy.
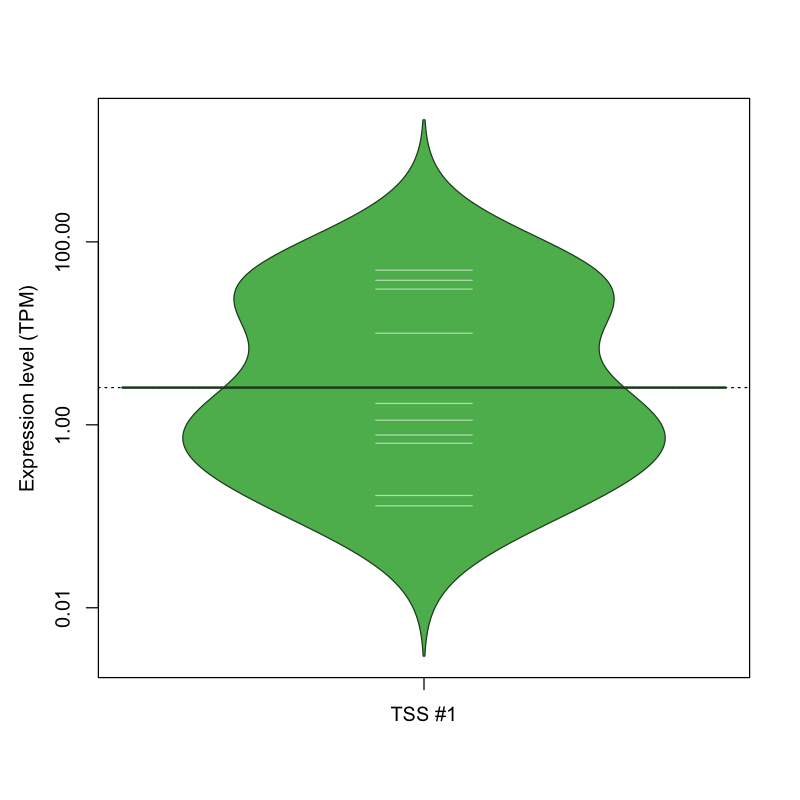
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_153203 | 1062 libraries | 4 libraries | 2 libraries | 0 libraries | 4 libraries |
The graphical summary, for retro_mmus_3356 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3356 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3356 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 8 parental genes, and 15 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000011042 | 2 retrocopies | |
| Equus caballus | ENSECAG00000014403 | 1 retrocopy | |
| Felis catus | ENSFCAG00000022091 | 2 retrocopies | |
| Monodelphis domestica | ENSMODG00000023336 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000003660 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000023004 | 6 retrocopies | |
| Mus musculus | ENSMUSG00000043091 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000067338 | 1 retrocopy |
retro_mmus_3356 ,
|