RetrogeneDB ID: | retro_hsap_2593 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 22:42503771..42504957(-) | ||
| Located in intron of: | ENSG00000237037 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000213790 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | OLA1 | ||
| Ensembl ID: | ENSG00000138430 | ||
| Aliases: | OLA1, DOC45, GBP45, GTBP9, GTPBP9 | ||
| Description: | Obg-like ATPase 1 [Source:HGNC Symbol;Acc:28833] |

Retrocopy-Parental alignment summary:
>retro_hsap_2593
ATGCCCCCTCAAAAGGGAGGTGATGGAATTAAACCACCCCCAATCATTGGAAGATTTGGAACCTCACTGAAAATTGGTAT
TGTTGGATTGCCAAATGTTGGGAAATCTACTTTCTTCAATGTATTAACCAATAGTCAGGCTTCAGCAGAAAACTTCCCAT
TCTGCACTATTGATCCTAATGAGAGCAGAGTACCTGTGCCAGATGAAAGGTTTGACTTTCTTTGCCAATATCACAAACCA
GCAAGCAAAATTCCTGCCTTTCTAAATGTAGTGGATATTGCTGGCCTTGTGAAAGGAGCTCACAATGGGCAGGGCCTGGG
GAATGCTTTTTTATCTCATTTTAGTGCTTGTGATGGCATCTTTCATCTAACACGTGCTTTTGAAGATGATGATATCACAC
ATGTTGAAGAAAGTGTAGATCCTATTCGAGATATAGAAATAATACATGAAGAGCTTCAGCTTAAAGATGAGGAATGACTG
GGCCCATTATAGATAAACTAGAAAAGGTGGCTGTGAGAGGAGGAGATAAAAAACTAAAACCCAAATATGATATAATGTGC
AAAGTAAAATCCTGGGTTATAGATCAAAAGAACCTGTTCGCTTCTATCATGATTGGAATGACAAAGAGATTGAAGTGTTG
AATAAACACTTATTTTTGACTTCAAAACCAATGGTCTACTTGGTTAATCTTTCTGAAAAAGACTACATTAGAAAGAAAAA
CAAATGGCTGATAAAAATTAAAGAGTGGGTGGACAAGTATGACCCAGGTGCCTTGGTCATTCCTTTTAGTGGGGCCTTGG
AACTCAAGTTGCAAGAATTGAGTGCTGAGGAGAGACAGCAGTATCTGGAAGCGAACATGACACAAAGTGCTTTGCCAAAG
ATCATTAAGGCTGGGTTTGCAGCACTCCAACTAAAATACTTTTTCACTGCAGGCCCAGATGAAGTGCGTGCACGGACCAT
CAGGAAAGGGACTAAGGCTCCTCAGGCTGCAGGAAAGATTCACACAGATTTTGAAAAGGGATTCATTATGGCTGAAGTAA
TGAAATATGAAGATTTTAAAGAGGAAGGTTCTGAAAATGCAGTCAAGGCTGCTGGAAAGTACAGACAACAAGGCAGAAAT
TATATTGTTGAAGATGGAGATATTATCTTCTTCAAATTTAACACACCTTAACAACTGAAGAAGAAA
ATGCCCCCTCAAAAGGGAGGTGATGGAATTAAACCACCCCCAATCATTGGAAGATTTGGAACCTCACTGAAAATTGGTAT
TGTTGGATTGCCAAATGTTGGGAAATCTACTTTCTTCAATGTATTAACCAATAGTCAGGCTTCAGCAGAAAACTTCCCAT
TCTGCACTATTGATCCTAATGAGAGCAGAGTACCTGTGCCAGATGAAAGGTTTGACTTTCTTTGCCAATATCACAAACCA
GCAAGCAAAATTCCTGCCTTTCTAAATGTAGTGGATATTGCTGGCCTTGTGAAAGGAGCTCACAATGGGCAGGGCCTGGG
GAATGCTTTTTTATCTCATTTTAGTGCTTGTGATGGCATCTTTCATCTAACACGTGCTTTTGAAGATGATGATATCACAC
ATGTTGAAGAAAGTGTAGATCCTATTCGAGATATAGAAATAATACATGAAGAGCTTCAGCTTAAAGATGAGGAATGACTG
GGCCCATTATAGATAAACTAGAAAAGGTGGCTGTGAGAGGAGGAGATAAAAAACTAAAACCCAAATATGATATAATGTGC
AAAGTAAAATCCTGGGTTATAGATCAAAAGAACCTGTTCGCTTCTATCATGATTGGAATGACAAAGAGATTGAAGTGTTG
AATAAACACTTATTTTTGACTTCAAAACCAATGGTCTACTTGGTTAATCTTTCTGAAAAAGACTACATTAGAAAGAAAAA
CAAATGGCTGATAAAAATTAAAGAGTGGGTGGACAAGTATGACCCAGGTGCCTTGGTCATTCCTTTTAGTGGGGCCTTGG
AACTCAAGTTGCAAGAATTGAGTGCTGAGGAGAGACAGCAGTATCTGGAAGCGAACATGACACAAAGTGCTTTGCCAAAG
ATCATTAAGGCTGGGTTTGCAGCACTCCAACTAAAATACTTTTTCACTGCAGGCCCAGATGAAGTGCGTGCACGGACCAT
CAGGAAAGGGACTAAGGCTCCTCAGGCTGCAGGAAAGATTCACACAGATTTTGAAAAGGGATTCATTATGGCTGAAGTAA
TGAAATATGAAGATTTTAAAGAGGAAGGTTCTGAAAATGCAGTCAAGGCTGCTGGAAAGTACAGACAACAAGGCAGAAAT
TATATTGTTGAAGATGGAGATATTATCTTCTTCAAATTTAACACACCTTAACAACTGAAGAAGAAA
ORF - retro_hsap_2593 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 96.73 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MPPKKGGDGIKPPPIIGRFGTSLKIGIVGLPNVGKSTFFNVLTNSQASAENFPFCTIDPNESRVPVPDER |
| MPP.KGGDGIKPPPIIGRFGTSLKIGIVGLPNVGKSTFFNVLTNSQASAENFPFCTIDPNESRVPVPDER | |
| Retrocopy | MPPQKGGDGIKPPPIIGRFGTSLKIGIVGLPNVGKSTFFNVLTNSQASAENFPFCTIDPNESRVPVPDER |
| Parental | FDFLCQYHKPASKIPAFLNVVDIAGLVKGAHNGQGLGNAFLSHISACDGIFHLTRAFEDDDITHVEGSVD |
| FDFLCQYHKPASKIPAFLNVVDIAGLVKGAHNGQGLGNAFLSH.SACDGIFHLTRAFEDDDITHVE.SVD | |
| Retrocopy | FDFLCQYHKPASKIPAFLNVVDIAGLVKGAHNGQGLGNAFLSHFSACDGIFHLTRAFEDDDITHVEESVD |
| Parental | PIRDIEIIHEELQLKDEE-MIGPIIDKLEKVAVRGGDKKLKPEYDIMCKVKSWVIDQK-KPVRFYHDWND |
| PIRDIEIIHEELQLKDEE.M.GPIIDKLEKVAVRGGDKKLKP.YDIMCKVKSWVIDQK..PVRFYHDWND | |
| Retrocopy | PIRDIEIIHEELQLKDEE<MTGPIIDKLEKVAVRGGDKKLKPKYDIMCKVKSWVIDQK<EPVRFYHDWND |
| Parental | KEIEVLNKHLFLTSKPMVYLVNLSEKDYIRKKNKWLIKIKEWVDKYDPGALVIPFSGALELKLQELSAEE |
| KEIEVLNKHLFLTSKPMVYLVNLSEKDYIRKKNKWLIKIKEWVDKYDPGALVIPFSGALELKLQELSAEE | |
| Retrocopy | KEIEVLNKHLFLTSKPMVYLVNLSEKDYIRKKNKWLIKIKEWVDKYDPGALVIPFSGALELKLQELSAEE |
| Parental | RQKYLEANMTQSALPKIIKAGFAALQLEYFFTAGPDEVRAWTIRKGTKAPQAAGKIHTDFEKGFIMAEVM |
| RQ.YLEANMTQSALPKIIKAGFAALQL.YFFTAGPDEVRA.TIRKGTKAPQAAGKIHTDFEKGFIMAEVM | |
| Retrocopy | RQQYLEANMTQSALPKIIKAGFAALQLKYFFTAGPDEVRARTIRKGTKAPQAAGKIHTDFEKGFIMAEVM |
| Parental | KYEDFKEEGSENAVKAAGKYRQQGRNYIVEDGDIIFFKFNTPQQPKKK |
| KYEDFKEEGSENAVKAAGKYRQQGRNYIVEDGDIIFFKFNTP.Q.KKK | |
| Retrocopy | KYEDFKEEGSENAVKAAGKYRQQGRNYIVEDGDIIFFKFNTP*QLKKK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .60 RPM | 59 .18 RPM |
| bodymap2_adrenal | 0 .49 RPM | 59 .57 RPM |
| bodymap2_brain | 0 .75 RPM | 133 .23 RPM |
| bodymap2_breast | 0 .49 RPM | 72 .59 RPM |
| bodymap2_colon | 0 .73 RPM | 87 .88 RPM |
| bodymap2_heart | 0 .59 RPM | 51 .82 RPM |
| bodymap2_kidney | 0 .35 RPM | 73 .99 RPM |
| bodymap2_liver | 0 .27 RPM | 34 .71 RPM |
| bodymap2_lung | 0 .60 RPM | 41 .77 RPM |
| bodymap2_lymph_node | 0 .69 RPM | 48 .96 RPM |
| bodymap2_ovary | 0 .79 RPM | 59 .76 RPM |
| bodymap2_prostate | 2 .34 RPM | 83 .02 RPM |
| bodymap2_skeletal_muscle | 1 .44 RPM | 104 .11 RPM |
| bodymap2_testis | 0 .89 RPM | 94 .27 RPM |
| bodymap2_thyroid | 0 .26 RPM | 75 .31 RPM |
| bodymap2_white_blood_cells | 1 .20 RPM | 65 .44 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_2593 was not detected
No EST(s) were mapped for retro_hsap_2593 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_109359 | 985 libraries | 763 libraries | 81 libraries | 0 libraries | 0 libraries |
The graphical summary, for retro_hsap_2593 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
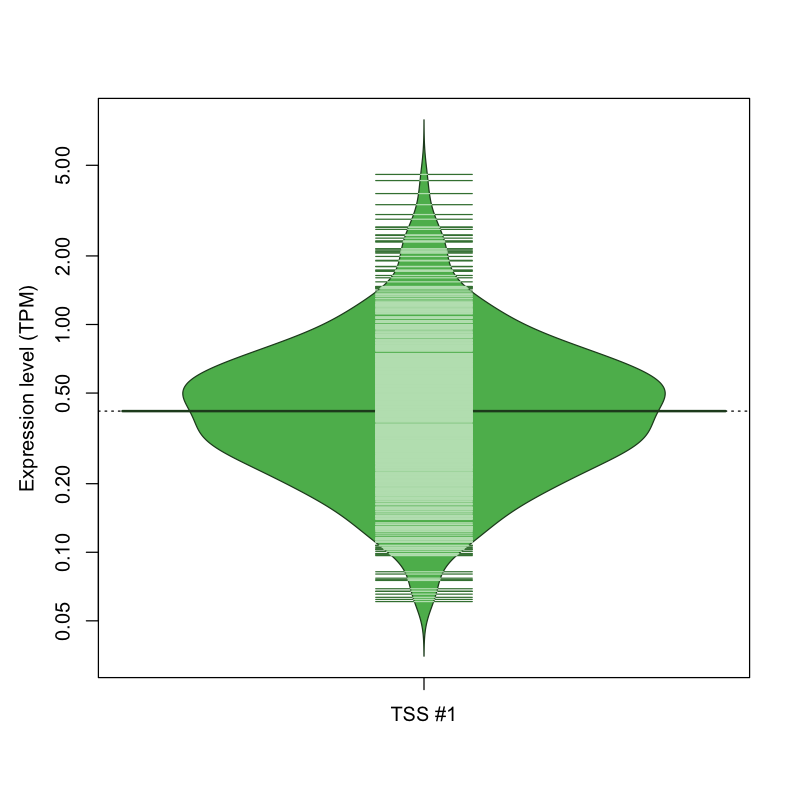
retro_hsap_2593 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2593 has 3 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_1517 |
| Gorilla gorilla | retro_ggor_1623 |
| Pongo abelii | retro_pabe_1912 |
Parental genes homology:
Parental genes homology involve 15 parental genes, and 18 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000006970 | 1 retrocopy | |
| Choloepus hoffmanni | ENSCHOG00000005044 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000007877 | 1 retrocopy | |
| Dipodomys ordii | ENSDORG00000008681 | 1 retrocopy | |
| Homo sapiens | ENSG00000138430 | 1 retrocopy |
retro_hsap_2593 ,
|
| Gorilla gorilla | ENSGGOG00000022215 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000025196 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000009296 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000005817 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000016557 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000012943 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000012650 | 1 retrocopy | |
| Sorex araneus | ENSSARG00000010743 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000005517 | 2 retrocopies | |
| Vicugna pacos | ENSVPAG00000009660 | 2 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .17 RPM |
| CEU_NA11843 | 0 .20 RPM |
| CEU_NA11930 | 0 .33 RPM |
| CEU_NA12004 | 0 .11 RPM |
| CEU_NA12400 | 0 .21 RPM |
| CEU_NA12751 | 0 .10 RPM |
| CEU_NA12760 | 0 .40 RPM |
| CEU_NA12827 | 0 .28 RPM |
| CEU_NA12872 | 0 .16 RPM |
| CEU_NA12873 | 0 .16 RPM |
| FIN_HG00183 | 0 .49 RPM |
| FIN_HG00277 | 0 .30 RPM |
| FIN_HG00315 | 0 .08 RPM |
| FIN_HG00321 | 0 .21 RPM |
| FIN_HG00328 | 0 .12 RPM |
| FIN_HG00338 | 0 .09 RPM |
| FIN_HG00349 | 0 .26 RPM |
| FIN_HG00375 | 0 .22 RPM |
| FIN_HG00377 | 0 .10 RPM |
| FIN_HG00378 | 0 .11 RPM |
| GBR_HG00099 | 0 .26 RPM |
| GBR_HG00111 | 0 .13 RPM |
| GBR_HG00114 | 0 .11 RPM |
| GBR_HG00119 | 0 .10 RPM |
| GBR_HG00131 | 0 .17 RPM |
| GBR_HG00133 | 0 .14 RPM |
| GBR_HG00134 | 0 .11 RPM |
| GBR_HG00137 | 0 .14 RPM |
| GBR_HG00142 | 0 .06 RPM |
| GBR_HG00143 | 0 .06 RPM |
| TSI_NA20512 | 0 .08 RPM |
| TSI_NA20513 | 0 .17 RPM |
| TSI_NA20518 | 0 .58 RPM |
| TSI_NA20532 | 0 .17 RPM |
| TSI_NA20538 | 0 .32 RPM |
| TSI_NA20756 | 0 .12 RPM |
| TSI_NA20765 | 0 .17 RPM |
| TSI_NA20771 | 0 .14 RPM |
| TSI_NA20786 | 0 .13 RPM |
| TSI_NA20798 | 0 .06 RPM |
| YRI_NA18870 | 0 .27 RPM |
| YRI_NA18907 | 0 .10 RPM |
| YRI_NA18916 | 0 .11 RPM |
| YRI_NA19093 | 0 .08 RPM |
| YRI_NA19099 | 0 .13 RPM |
| YRI_NA19114 | 0 .18 RPM |
| YRI_NA19118 | 0 .17 RPM |
| YRI_NA19213 | 0 .14 RPM |
| YRI_NA19214 | 0 .05 RPM |
| YRI_NA19223 | 0 .06 RPM |