RetrogeneDB ID: | retro_mmus_10 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 8:67514231..67515590(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000030735 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tufm | ||
| Ensembl ID: | ENSMUSG00000073838 | ||
| Aliases: | Tufm, 2300002G02Rik, C76308, C76389, EF-TuMT, EFTU | ||
| Description: | Tu translation elongation factor, mitochondrial [Source:MGI Symbol;Acc:MGI:1923686] |

Retrocopy-Parental alignment summary:
>retro_mmus_10
ATGGCGGCAGCCACTCTATTGCGAGCGACGCCCCGCTTCAGCGGTCTCTGCGCCAGCCCGACCCCATTTCTGCAAGGTCG
GCTGCGGCCGCTGAAGGCTCCCGCATCACCCTTCTTGTGTCGCGGCCTAGCTGTGGAGGCCAAGAAGACTTATGTACGCG
ACAAGCCCCATGTGAATGTGGGTACCATCGGACATGTGGACCATGGCAAGACCACCCTGACTGCAGCCATCACGAAAATT
CTAGCCGAGGGAGGTGGAGCTAAGTTCAAGAAGTACGAGGAGATAGACAATGCCCCAGAGGAGCGAGCTCGGGGCATCAC
CATCAATGCAGCCCATGTGGAGTATAGCACTGCTGCCCGACACTATGCCCACACAGACTGTCCTGGTCATGCAGATTATG
TTAAGAATATGATCACAGGCACTGCCCCCCTGGATGGCTGTATCCTGGTGGTGGCAGCTAATGATAGCCCCATGCCCCAG
ACCCGAGAGCACTTACTGCTAGCTAAACAGATTGGGGTAGAACATGTTGTGGTGTACGTGAACAAGGCAGACGCTGTCCA
GGACTCAGAGATGGTGGAGTTAGTCGAGCTGGAGATCCGGGAGCTGCTCACCGAGTTTGGCTATAAAGGAGAGGAAACTC
CAGTCATTGTAGGCTCGGCTCTCTGTGCCCTTGAGCAACGTGACCCTGAGCTAGGCGTGAAGTCAGTGCAGAAGCTCCTG
GATGCTGTGGACACCTACATCCCAGTGCCCACCCGGGACCTGGACAAGCCCTTCCTGCTCCCTGTAGAGTCAGTCTACTC
TATTCCTGGCCGGGGCACAGTGGTGACAGGTACATTAGAGCGTGGCATTTTGAAGAAAGGAGATGAGTGTGAGTTGCTGG
GACATAACAAGAACATCCGCACTGTGGTGACAGGCATTGAGATGTTCCACAAGAGCCTGGAGAGGGCTGAGGCAGGGGAT
AACCTGGGTGCTCTGGTCCGAGGCTTAAAGCGCGAAGATTTGAGGCGTGGCTTGGTCATGGTCAAGCCAGGCTCCATCCA
ACCCCACCAGAAGGTGGAGGCCCAGGTTTATATCCTCAGCAAGGAGGAAGGTGGCCGCCACAAACCCTTTGTATCTCATT
TCATGCCCGTCATGTTCTCCCTGACCTGGGACATGGCCTGTCGAGTCATCTTGCCTCCAGGGAAGGAACTTGCCATGCCT
GGAGAGGACTTGAAGCTTAGTCTAATCTTGCGGCAGCCCATGATCTTAGAGAAAGGCCAACGTTTCACCTTGAGGGATGG
CAACAAGACCATTGGCACTGGCCTTGTTACTGATGTACCAGCCATGACTGAGGAGGACAAGAACATCAAGTGGAGCTGA
ATGGCGGCAGCCACTCTATTGCGAGCGACGCCCCGCTTCAGCGGTCTCTGCGCCAGCCCGACCCCATTTCTGCAAGGTCG
GCTGCGGCCGCTGAAGGCTCCCGCATCACCCTTCTTGTGTCGCGGCCTAGCTGTGGAGGCCAAGAAGACTTATGTACGCG
ACAAGCCCCATGTGAATGTGGGTACCATCGGACATGTGGACCATGGCAAGACCACCCTGACTGCAGCCATCACGAAAATT
CTAGCCGAGGGAGGTGGAGCTAAGTTCAAGAAGTACGAGGAGATAGACAATGCCCCAGAGGAGCGAGCTCGGGGCATCAC
CATCAATGCAGCCCATGTGGAGTATAGCACTGCTGCCCGACACTATGCCCACACAGACTGTCCTGGTCATGCAGATTATG
TTAAGAATATGATCACAGGCACTGCCCCCCTGGATGGCTGTATCCTGGTGGTGGCAGCTAATGATAGCCCCATGCCCCAG
ACCCGAGAGCACTTACTGCTAGCTAAACAGATTGGGGTAGAACATGTTGTGGTGTACGTGAACAAGGCAGACGCTGTCCA
GGACTCAGAGATGGTGGAGTTAGTCGAGCTGGAGATCCGGGAGCTGCTCACCGAGTTTGGCTATAAAGGAGAGGAAACTC
CAGTCATTGTAGGCTCGGCTCTCTGTGCCCTTGAGCAACGTGACCCTGAGCTAGGCGTGAAGTCAGTGCAGAAGCTCCTG
GATGCTGTGGACACCTACATCCCAGTGCCCACCCGGGACCTGGACAAGCCCTTCCTGCTCCCTGTAGAGTCAGTCTACTC
TATTCCTGGCCGGGGCACAGTGGTGACAGGTACATTAGAGCGTGGCATTTTGAAGAAAGGAGATGAGTGTGAGTTGCTGG
GACATAACAAGAACATCCGCACTGTGGTGACAGGCATTGAGATGTTCCACAAGAGCCTGGAGAGGGCTGAGGCAGGGGAT
AACCTGGGTGCTCTGGTCCGAGGCTTAAAGCGCGAAGATTTGAGGCGTGGCTTGGTCATGGTCAAGCCAGGCTCCATCCA
ACCCCACCAGAAGGTGGAGGCCCAGGTTTATATCCTCAGCAAGGAGGAAGGTGGCCGCCACAAACCCTTTGTATCTCATT
TCATGCCCGTCATGTTCTCCCTGACCTGGGACATGGCCTGTCGAGTCATCTTGCCTCCAGGGAAGGAACTTGCCATGCCT
GGAGAGGACTTGAAGCTTAGTCTAATCTTGCGGCAGCCCATGATCTTAGAGAAAGGCCAACGTTTCACCTTGAGGGATGG
CAACAAGACCATTGGCACTGGCCTTGTTACTGATGTACCAGCCATGACTGAGGAGGACAAGAACATCAAGTGGAGCTGA
ORF - retro_mmus_10 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 99.78 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAAATLLRATPRFSGLCASPTPFLQGRLRPLKAPASPFLCRGLAVEAKKTYVRDKPHVNVGTIGHVDHGK |
| MAAATLLRATPRFSGLCASPTPFLQGRLRPLKAPASPFLCRGLAVEAKKTYVRDKPHVNVGTIGHVDHGK | |
| Retrocopy | MAAATLLRATPRFSGLCASPTPFLQGRLRPLKAPASPFLCRGLAVEAKKTYVRDKPHVNVGTIGHVDHGK |
| Parental | TTLTAAITKILAEGGGAKFKKYEEIDNAPEERARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITG |
| TTLTAAITKILAEGGGAKFKKYEEIDNAPEERARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITG | |
| Retrocopy | TTLTAAITKILAEGGGAKFKKYEEIDNAPEERARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITG |
| Parental | TAPLDGCILVVAANDGPMPQTREHLLLAKQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKG |
| TAPLDGCILVVAAND.PMPQTREHLLLAKQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKG | |
| Retrocopy | TAPLDGCILVVAANDSPMPQTREHLLLAKQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKG |
| Parental | EETPVIVGSALCALEQRDPELGVKSVQKLLDAVDTYIPVPTRDLDKPFLLPVESVYSIPGRGTVVTGTLE |
| EETPVIVGSALCALEQRDPELGVKSVQKLLDAVDTYIPVPTRDLDKPFLLPVESVYSIPGRGTVVTGTLE | |
| Retrocopy | EETPVIVGSALCALEQRDPELGVKSVQKLLDAVDTYIPVPTRDLDKPFLLPVESVYSIPGRGTVVTGTLE |
| Parental | RGILKKGDECELLGHNKNIRTVVTGIEMFHKSLERAEAGDNLGALVRGLKREDLRRGLVMVKPGSIQPHQ |
| RGILKKGDECELLGHNKNIRTVVTGIEMFHKSLERAEAGDNLGALVRGLKREDLRRGLVMVKPGSIQPHQ | |
| Retrocopy | RGILKKGDECELLGHNKNIRTVVTGIEMFHKSLERAEAGDNLGALVRGLKREDLRRGLVMVKPGSIQPHQ |
| Parental | KVEAQVYILSKEEGGRHKPFVSHFMPVMFSLTWDMACRVILPPGKELAMPGEDLKLSLILRQPMILEKGQ |
| KVEAQVYILSKEEGGRHKPFVSHFMPVMFSLTWDMACRVILPPGKELAMPGEDLKLSLILRQPMILEKGQ | |
| Retrocopy | KVEAQVYILSKEEGGRHKPFVSHFMPVMFSLTWDMACRVILPPGKELAMPGEDLKLSLILRQPMILEKGQ |
| Parental | RFTLRDGNKTIGTGLVTDVPAMTEEDKNIKWS |
| RFTLRDGNKTIGTGLVTDVPAMTEEDKNIKWS | |
| Retrocopy | RFTLRDGNKTIGTGLVTDVPAMTEEDKNIKWS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .21 RPM | 16 .84 RPM |
| SRP007412_cerebellum | 0 .22 RPM | 20 .76 RPM |
| SRP007412_heart | 0 .87 RPM | 81 .00 RPM |
| SRP007412_kidney | 0 .65 RPM | 58 .15 RPM |
| SRP007412_liver | 0 .25 RPM | 38 .53 RPM |
| SRP007412_testis | 0 .09 RPM | 16 .74 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_10 was not detected
No EST(s) were mapped for retro_mmus_10 retrocopy.
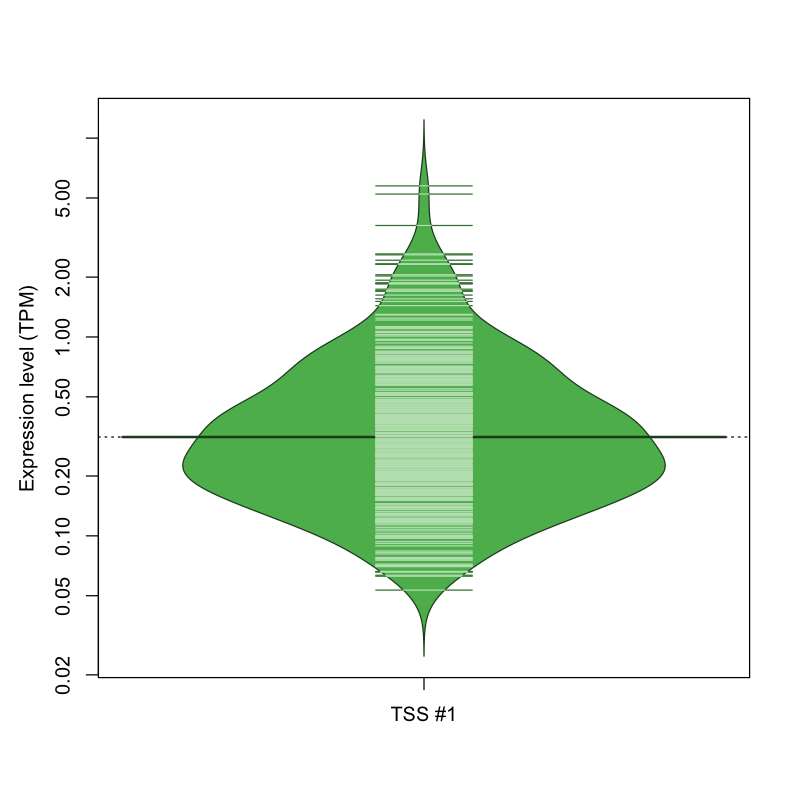
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_143140 | 685 libraries | 352 libraries | 33 libraries | 2 libraries | 0 libraries |
The graphical summary, for retro_mmus_10 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_10 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_10 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 11 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Dipodomys ordii | ENSDORG00000016474 | 1 retrocopy | |
| Homo sapiens | ENSG00000178952 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000011131 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000015564 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000073838 | 1 retrocopy |
retro_mmus_10 ,
|
| Ornithorhynchus anatinus | ENSOANG00000013114 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000002683 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000007939 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000018604 | 1 retrocopy | |
| Vicugna pacos | ENSVPAG00000009541 | 1 retrocopy | |
| Drosophila melanogaster | FBgn0033184 | 1 retrocopy |