RetrogeneDB ID: | retro_mmus_1372 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 15:93412747..93413283(+) | ||
| Located in intron of: | ENSMUSG00000036167 | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Rac1 | ||
| Ensembl ID: | ENSMUSG00000001847 | ||
| Aliases: | Rac1, AL023026, D5Ertd559e | ||
| Description: | RAS-related C3 botulinum substrate 1 [Source:MGI Symbol;Acc:MGI:97845] |

Retrocopy-Parental alignment summary:
>retro_mmus_1372
GCTGCTGGTAAAACCTGCCTGCTCATCAGTTACCCGACCAATGCATTTCCTGGACAGTACATCCCCACCGTCTTTGACAA
CTAGTCTGCCAATGTTATGGTAGATGGAAAACCAGTGAATCTGGGCCTATGGGACACAGCTGGACAAGAAGATCATGACA
GATTGCATCCCCTCTCCTACCTGCAAACAGACGTGTTCTTAATTTGCTTTTCCCTTGTGAGTCCTGCATCATTTGAAAAT
GTCTGTGTAAAGTGGTCTCCTGAAGTGCGACACCACTGTCCCAATACTCCTATCATCCTAGTGGGGACGAAGCTTGATCT
TAGGGATGATAAGGACACCATTGAGAAGCTGAAGGAGAAGAAGCTGACTCCCATCACCTACCCGCAGGGGTAGCCATGGC
CAAAGAGATCGGTGCTGTTAAATACCTGGAGTGCTCAGCTCTCACACAGCAAGTACTCAAGACAGTGTTTGACGAAGCTA
TCCGAGCAGTTCTCTGTCCCCCTCCTGTCAAGAAGAGGAGAGAATGCCTGCTGTTG
GCTGCTGGTAAAACCTGCCTGCTCATCAGTTACCCGACCAATGCATTTCCTGGACAGTACATCCCCACCGTCTTTGACAA
CTAGTCTGCCAATGTTATGGTAGATGGAAAACCAGTGAATCTGGGCCTATGGGACACAGCTGGACAAGAAGATCATGACA
GATTGCATCCCCTCTCCTACCTGCAAACAGACGTGTTCTTAATTTGCTTTTCCCTTGTGAGTCCTGCATCATTTGAAAAT
GTCTGTGTAAAGTGGTCTCCTGAAGTGCGACACCACTGTCCCAATACTCCTATCATCCTAGTGGGGACGAAGCTTGATCT
TAGGGATGATAAGGACACCATTGAGAAGCTGAAGGAGAAGAAGCTGACTCCCATCACCTACCCGCAGGGGTAGCCATGGC
CAAAGAGATCGGTGCTGTTAAATACCTGGAGTGCTCAGCTCTCACACAGCAAGTACTCAAGACAGTGTTTGACGAAGCTA
TCCGAGCAGTTCTCTGTCCCCCTCCTGTCAAGAAGAGGAGAGAATGCCTGCTGTTG
ORF - retro_mmus_1372 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 91.16 % |
| Parental protein coverage: | 93.75 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | AVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDGKPVNLGLWDTAGQEDYDRLRPLSYPQTDVFLICF |
| A.GKTCLLISY.TNAFPG.YIPTVFDN.SANVMVDGKPVNLGLWDTAGQED.DRL.PLSY.QTDVFLICF | |
| Retrocopy | AAGKTCLLISYPTNAFPGQYIPTVFDN*SANVMVDGKPVNLGLWDTAGQEDHDRLHPLSYLQTDVFLICF |
| Parental | SLVSPASFENVRAKWYPEVRHHCPNTPIILVGTKLDLRDDKDTIEKLKEKKLTPITYPQG-LAMAKEIGA |
| SLVSPASFENV..KW.PEVRHHCPNTPIILVGTKLDLRDDKDTIEKLKEKKLTPITYPQG..AMAKEIGA | |
| Retrocopy | SLVSPASFENVCVKWSPEVRHHCPNTPIILVGTKLDLRDDKDTIEKLKEKKLTPITYPQG<VAMAKEIGA |
| Parental | VKYLECSALTQRGLKTVFDEAIRAVLCPPPVKKRKRKCLLL |
| VKYLECSALTQ..LKTVFDEAIRAVLCPPPVKKR.R.CLLL | |
| Retrocopy | VKYLECSALTQQVLKTVFDEAIRAVLCPPPVKKR-RECLLL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .16 RPM | 233 .30 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 180 .20 RPM |
| SRP007412_heart | 0 .03 RPM | 117 .67 RPM |
| SRP007412_kidney | 0 .06 RPM | 141 .31 RPM |
| SRP007412_liver | 0 .14 RPM | 97 .08 RPM |
| SRP007412_testis | 0 .00 RPM | 23 .88 RPM |
RNA Polymerase II actvity may be related with retro_mmus_1372 in 1 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001YKA | POLR2A | 15:93411248..93412167 |
| ENCFF001YKA | POLR2A | 15:93412617..93412953 |
No EST(s) were mapped for retro_mmus_1372 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
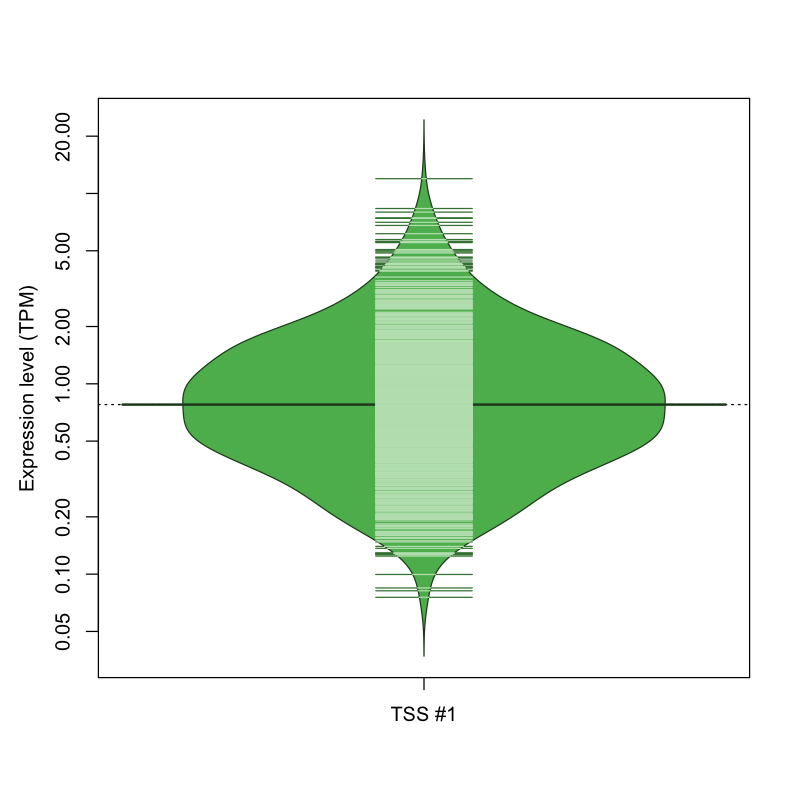
| TSS #1 | TSS_43851 | 248 libraries | 505 libraries | 305 libraries | 13 libraries | 1 library |
The graphical summary, for retro_mmus_1372 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1372 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1372 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 10 parental genes, and 26 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000136238 | 6 retrocopies | |
| Gorilla gorilla | ENSGGOG00000026733 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000001847 | 2 retrocopies |
retro_mmus_1372 , retro_mmus_234,
|
| Mus musculus | ENSMUSG00000006699 | 3 retrocopies | |
| Pan troglodytes | ENSPTRG00000018910 | 4 retrocopies | |
| Rattus norvegicus | ENSRNOG00000001068 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000006784 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000007272 | 4 retrocopies | |
| Tarsius syrichta | ENSTSYG00000001618 | 2 retrocopies | |
| Xenopus tropicalis | ENSXETG00000027593 | 1 retrocopy |