RetrogeneDB ID: | retro_mmus_3229 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 8:97427714..97429110(-) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000081111 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | G3bp1 | ||
| Ensembl ID: | ENSMUSG00000018583 | ||
| Aliases: | None | ||
| Description: | GTPase activating protein (SH3 domain) binding protein 1 [Source:MGI Symbol;Acc:MGI:1351465] |

Retrocopy-Parental alignment summary:
>retro_mmus_3229
ATGGTTATGGAGAAGCCTAGTCCCCTGCTGGTCAGGCGGGAGTTTGTGCGACAGTACTACACCCTGCTGAACCAGGCCCT
GGACATGCTGCACAGATTTTATGGAAAGAACTCTTCCTATGCCCACGGGGGCTTGGATTCCAACGGGAAGCCAGCAGATG
CAGTCTACGGGAAGAAGGAAATCCACAGGAAAGTGATGTCACAAAACTTCACCAACTGCCACACCAAGATCTGCCACGTG
GATGCTCACGCAACTCTGAATGACGGGGTGGTTGTCCAAGTGATGGGGCTACTGTCCAACAACAACCAGGCTCTGCGGAG
GTTCATGCAGACCTTTGTCCTTGCTCCTGAGGGCTCTGCTGCCAACAAATTCTATGTTCACAACGACATCTTCAGATACC
AAGATGAGGTCTTCGGTGGCTTTGTCACAGAGCCTCAAAAGGAATCCGAGGAAGAAGTAGAAGAACCTGAAGAAAGACAG
CAGACACCAGAGGTGGTGCCTGATGATTCTGGAACTTTCTATGATCAGACTGTCAGCAATGACTTGGAGGAGCATTTAGA
GGAGCCTGTAGTGGAACCAGAACCGGAGCCAGAGCCGGAGCCAGAGCCGGAACCTGTGTCCAACATTCAAGAGGACAAGC
CTGAGGCTGCATTGGAAGAAGCTGCTCCCGACGATGTGCAGAAGAGCACTTCCCCGACCCCGGCTGACGTGGCCCCGGCA
CAGGAGGACTTGAGGACGTTTTCTTGGACATCTATGACGAGTAAGAACCTTCCTCCCAGTGGGGCTGTTCCAGTGACGGG
GACATCACCTCATGTGGTCAAAGTGCCAGCGTCACAGCCCGGTCCAGAATCTAAACCTGACTCTCAGATCCCACCACAAA
GGCCTCAGAGAGATCAGAGAGTTCGAGAGCAGCGAATCAATATCCCTCCTCAGAGAGGACCCCGACCAATCCGTGAGGCT
GGTGAACCAGGAGATGTGGAACCTCGAAGGATGGTGAGGCACCCTGACAGTCACCAGCTCTTCACTGGAAACCTGCCACA
TGAAGTTGACAAGTCGGAACTGAAGGATTTTTTTCCAAAATTTTGGCAATGTGGTGGAGCTGCGCATCAACAGTGGTGGG
AAGTTACCCAATTTCGGTTTTGTTGTGTTTGATGATTCTGAACCTGTTCAGAAGGTCCTTAATAATAGGCCCATCATGTT
CCGAGGCGCGGTCCGCCTGAATGTGGAAGAGAAGAAGACTCGAGCTGCCAGAGAAGGCGACCGCAGGGATAACCGCCTTC
AGGGACCTGGAGGCCCCCGTGGTGGGCCGAGTGGTGGGATGAGAGGTCCTTCCCGTGGAGGCATGGTGCAGAAACCAGGC
TTTGGCGTGGGCAGGGGAATTACAACTCCAAGGCAG
ORF - retro_mmus_3229 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 92.06 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MVMEKPSPLLVGREFVRQYYTLLNQAPDMLHRFYGKNSSYAHGGLDSNGKPADAVYGQKEIHRKVMSQNF |
| MVMEKPSPLLV.REFVRQYYTLLNQA.DMLHRFYGKNSSYAHGGLDSNGKPADAVYG.KEIHRKVMSQNF | |
| Retrocopy | MVMEKPSPLLVRREFVRQYYTLLNQALDMLHRFYGKNSSYAHGGLDSNGKPADAVYGKKEIHRKVMSQNF |
| Parental | TNCHTKIRHVDAHATLNDGVVVQVMGLLSNNNQALRRFMQTFVLAPEGSVANKFYVHNDIFRYQDEVFGG |
| TNCHTKI.HVDAHATLNDGVVVQVMGLLSNNNQALRRFMQTFVLAPEGS.ANKFYVHNDIFRYQDEVFGG | |
| Retrocopy | TNCHTKICHVDAHATLNDGVVVQVMGLLSNNNQALRRFMQTFVLAPEGSAANKFYVHNDIFRYQDEVFGG |
| Parental | FVTEPQEESEEEVEEPEERQQTPEVVPDDSGTFYDQTVSNDLEEHLEEPVVEPEPEPEPEPEPEPVSDIQ |
| FVTEPQ.ESEEEVEEPEERQQTPEVVPDDSGTFYDQTVSNDLEEHL....................S.IQ | |
| Retrocopy | FVTEPQKESEEEVEEPEERQQTPEVVPDDSGTFYDQTVSNDLEEHLXXXXXXXXXXXXXXXXXXXXSNIQ |
| Parental | EDKPEAALEEAAPDDVQKSTSPAPADVAPAQEDLRTFSWASVTSKNLPPSGAVPVTGTPPHVVKVPASQP |
| EDKPEAALEEAAPDDVQKSTSP.PADVAPAQEDLRTFSW.S.TSKNLPPSGAVPVTGT.PHVVKVPASQP | |
| Retrocopy | EDKPEAALEEAAPDDVQKSTSPTPADVAPAQEDLRTFSWTSMTSKNLPPSGAVPVTGTSPHVVKVPASQP |
| Parental | RPESKPDSQIPPQRPQRDQRVREQRINIPPQRGPRPIREAGEPGDVEPRRMVRHPDSHQLFIGNLPHEVD |
| .PESKPDSQIPPQRPQRDQRVREQRINIPPQRGPRPIREAGEPGDVEPRRMVRHPDSHQLF.GNLPHEVD | |
| Retrocopy | GPESKPDSQIPPQRPQRDQRVREQRINIPPQRGPRPIREAGEPGDVEPRRMVRHPDSHQLFTGNLPHEVD |
| Parental | KSELKD-FFQNFGNVVELRINSGGKLPNFGFVVFDDSEPVQKVLSNRPIMFRGAVRLNVEEKKTRAAREG |
| KSELKD.FFQNFGNVVELRINSGGKLPNFGFVVFDDSEPVQKVL.NRPIMFRGAVRLNVEEKKTRAAREG | |
| Retrocopy | KSELKD>FFQNFGNVVELRINSGGKLPNFGFVVFDDSEPVQKVLNNRPIMFRGAVRLNVEEKKTRAAREG |
| Parental | DRRDNRLRGPGGPRGGPSGGMRGPPRGGMVQKPGFGVGRGITTPRQ |
| DRRDNRL.GPGGPRGGPSGGMRGP.RGGMVQKPGFGVGRGITTPRQ | |
| Retrocopy | DRRDNRLQGPGGPRGGPSGGMRGPSRGGMVQKPGFGVGRGITTPRQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 58 .22 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 51 .45 RPM |
| SRP007412_heart | 0 .47 RPM | 92 .49 RPM |
| SRP007412_kidney | 0 .21 RPM | 84 .14 RPM |
| SRP007412_liver | 0 .08 RPM | 51 .42 RPM |
| SRP007412_testis | 4 .62 RPM | 40 .66 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_3229 was not detected
No EST(s) were mapped for retro_mmus_3229 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
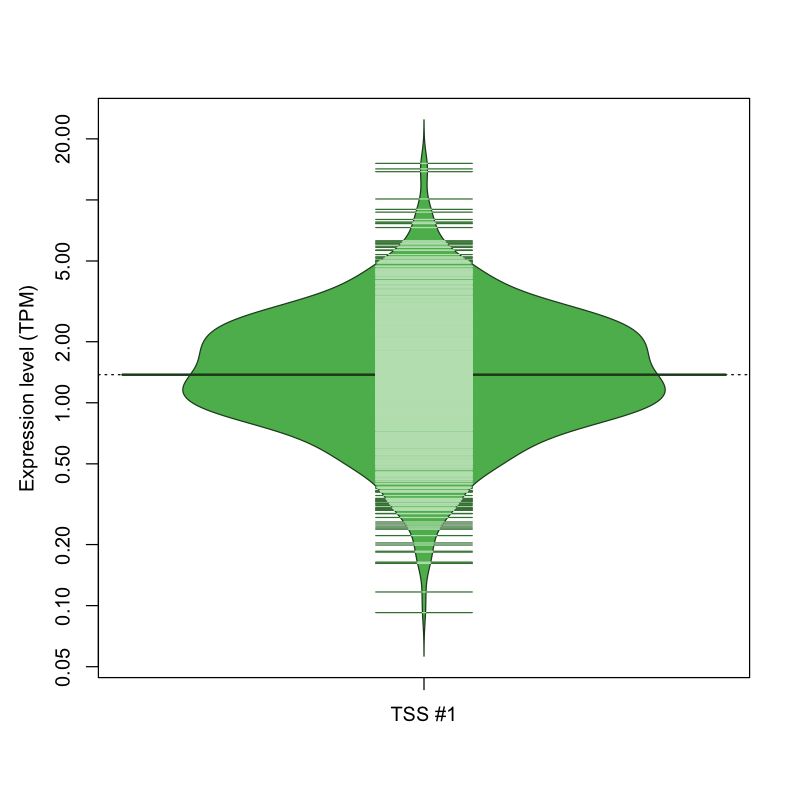
| TSS #1 | TSS_145737 | 122 libraries | 298 libraries | 626 libraries | 22 libraries | 4 libraries |
The graphical summary, for retro_mmus_3229 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3229 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3229 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 6 parental genes, and 10 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Cavia porcellus | ENSCPOG00000006626 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000018583 | 3 retrocopies |
retro_mmus_1389, retro_mmus_3229 , retro_mmus_3688,
|
| Nomascus leucogenys | ENSNLEG00000016470 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000022143 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000001726 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000013186 | 1 retrocopy |