RetrogeneDB ID: | retro_mmus_358 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:119526510..119527560(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000037015 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | BC023829 | ||
| Ensembl ID: | ENSMUSG00000073139 | ||
| Aliases: | BC023829, Tmem185a | ||
| Description: | cDNA sequence BC023829 [Source:MGI Symbol;Acc:MGI:2448555] |

Retrocopy-Parental alignment summary:
>retro_mmus_358
ATGAACCCCAGGGGCCTGTTCCAGGACTTCAACCCCAGTAAGTTCCTCATTTATGCCTGCTTGCTACTCTTCTCCGTGCT
GCTGCCCCTGCGCCTGGACGGCATCATCCAGTGGAGCTACTGGGCAGTCTTCGCCCCCATCTGGCTGTGGAAGCTCCTGG
TCATCGTGGGCGCCTCGGTGGGTGCGGGCGTGTGGGCCCGCAACCCACGCTACCGTACAGAGGGGGAAGCCTGCGTGGAA
TTCAAAGCCATGCTTATCGCCGTGGGCATCCACCTGCTACTACTCATGTTTGAGATACTAGTCTGCGACAGGGTGGAGAG
GGGCACCCACTTTTGGCTGCTGGTCTTCATGCCACTCTTCTTCGTTTCCCCTGTGTCCGTGGCCGCCTGTGTCTGGGGCT
TCCGACACGACAGGTCTTTGGAACTGGAGATCCTGTGCTCTGTCAACATCCTGCAGTTCATCTTCATCGCCCTGAGGCTG
GACAGGATTATCCACTGGCCGTGGCTAGTGGTGTTCGTGCCCCTGTGGATCCTCATGTCATTCCTCTGCCTGGTAGTTCT
CTATTACATCGTGTGGTCCCTCCTATTCCTGCGGTCCCTGGATGTGGTTGCAGAGCAGCGAAGAACACATGTGACCATGG
CCATCAGCTGGATCACCATTGTCGTGCCCCTGCTCATTTTTGAGGTCCTGCTAGTTCACAGACTGGATGACCACAATACG
TTCTCTTACATTTCCATTTTCATCCCGCTTTGGCTCTCATTACTGACTTTGATGGCTACAACGTTCAGGCGGAAAGGGGG
CAATCATTGGTGGTTTGGGATTCGCAGGGATTTCTGTCAGTTTCTACTAGAAGTTTTTCCGTTTTTAAGAGAATATGGAA
ATATCTCTTACGATCTCCATCACGAAGACAGTGAAGATGCTGAAGACGCATCGGTGTCAGAAGCTCCGAAAATTGCTCCA
ATGTTTGGAAAGAAGGCGCGGGTAGTTATAACACAGAGCCCTGGGAAATATGTGCCTCCACCTCCCAAGTTAAACATTGA
TATGCCAGAC
ORF - retro_mmus_358 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 88.29 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MNLRGLFQDFNPSKFLIYACLLLFSVLLALRLDGIIQWSYWAVFAPIWLWKLMVIVGASVGTGVWARNPQ |
| MN.RGLFQDFNPSKFLIYACLLLFSVLL.LRLDGIIQWSYWAVFAPIWLWKL.VIVGASVG.GVWARNP. | |
| Retrocopy | MNPRGLFQDFNPSKFLIYACLLLFSVLLPLRLDGIIQWSYWAVFAPIWLWKLLVIVGASVGAGVWARNPR |
| Parental | YRAEGETCVEFKAMLIAVGIHLLLLMFEVLVCDRIERGSHFWLLVFMPLFFVSPVSVAACVWGFRHDRSL |
| YR.EGE.CVEFKAMLIAVGIHLLLLMFE.LVCDR.ERG.HFWLLVFMPLFFVSPVSVAACVWGFRHDRSL | |
| Retrocopy | YRTEGEACVEFKAMLIAVGIHLLLLMFEILVCDRVERGTHFWLLVFMPLFFVSPVSVAACVWGFRHDRSL |
| Parental | ELEILCSVNILQFIFIALRLDKIIHWPWLVVCVPLWILMSFLCLVVLYYIVWSVLFLRSMDVIAEQRRTH |
| ELEILCSVNILQFIFIALRLD.IIHWPWLVV.VPLWILMSFLCLVVLYYIVWS.LFLRS.DV.AEQRRTH | |
| Retrocopy | ELEILCSVNILQFIFIALRLDRIIHWPWLVVFVPLWILMSFLCLVVLYYIVWSLLFLRSLDVVAEQRRTH |
| Parental | ITMALSWMTIVVPLLTFEILLVHKLDGHNAFSCIPIFVPLWLSLITLMATTFGQKGGNHWWFGIRKDFCQ |
| .TMA.SW.TIVVPLL.FE.LLVH.LD.HN.FS.I.IF.PLWLSL.TLMATTF..KGGNHWWFGIR.DFCQ | |
| Retrocopy | VTMAISWITIVVPLLIFEVLLVHRLDDHNTFSYISIFIPLWLSLLTLMATTFRRKGGNHWWFGIRRDFCQ |
| Parental | FLLEIFPFLREYGNISYDLHHEDSEETEETPVPEPPKIAPMFRKKARVVITQSPGKYVLPPPKLNIEMPD |
| FLLE.FPFLREYGNISYDLHHEDSE..E...V.E.PKIAPMF.KKARVVITQSPGKYV.PPPKLNI.MPD | |
| Retrocopy | FLLEVFPFLREYGNISYDLHHEDSEDAEDASVSEAPKIAPMFGKKARVVITQSPGKYVPPPPKLNIDMPD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 6 .45 RPM | 34 .48 RPM |
| SRP007412_cerebellum | 6 .64 RPM | 31 .31 RPM |
| SRP007412_heart | 6 .97 RPM | 16 .72 RPM |
| SRP007412_kidney | 12 .32 RPM | 22 .58 RPM |
| SRP007412_liver | 5 .16 RPM | 21 .03 RPM |
| SRP007412_testis | 6 .86 RPM | 13 .58 RPM |
RNA Polymerase II activity may be related with retro_mmus_358 in 3 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 1:119525846..119526680 |
| ENCFF001YIJ | POLR2A | 1:119525842..119526794 |
| ENCFF001YKA | POLR2A | 1:119525160..119530127 |
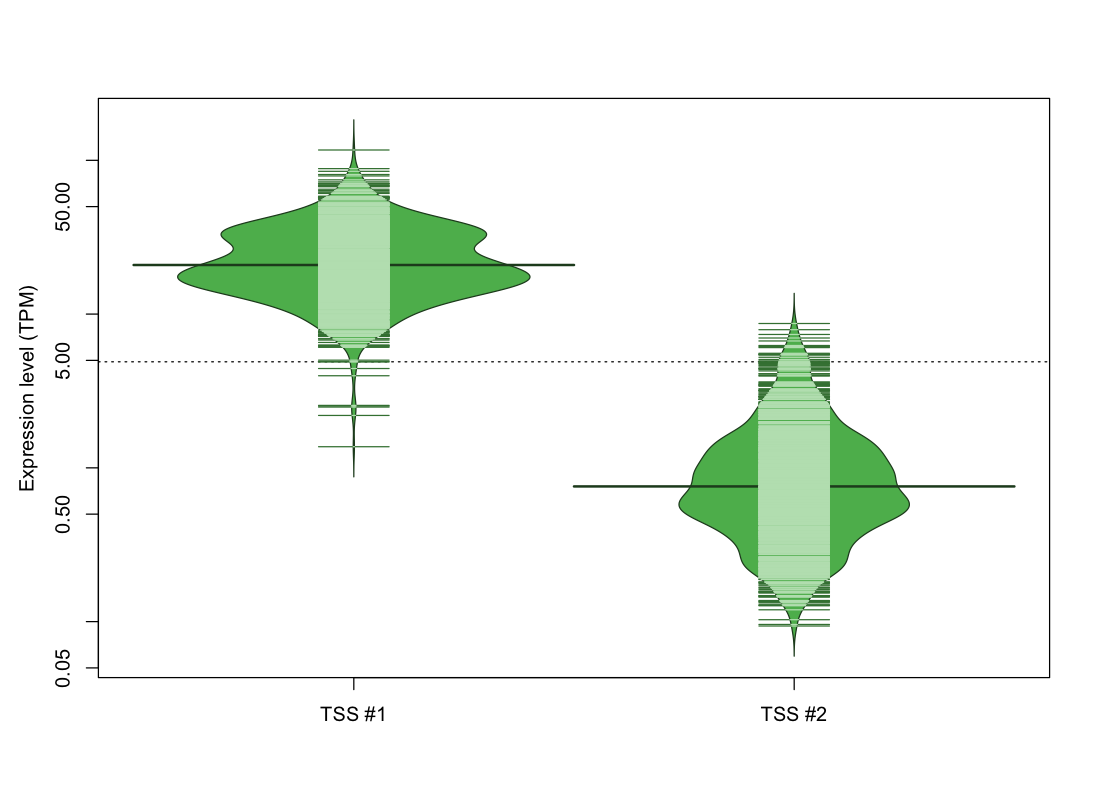
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_68244 | 10 libraries | 0 libraries | 9 libraries | 74 libraries | 979 libraries |
| TSS #2 | TSS_68245 | 247 libraries | 529 libraries | 281 libraries | 15 libraries | 0 libraries |
The graphical summary, for retro_mmus_358 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_358 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_358 has 5 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Homo sapiens | retro_hsap_104 |
| Gorilla gorilla | retro_ggor_189 |
| Pongo abelii | retro_pabe_118 |
| Macaca mulatta | retro_mmul_310 |
| Rattus norvegicus | retro_rnor_6 |