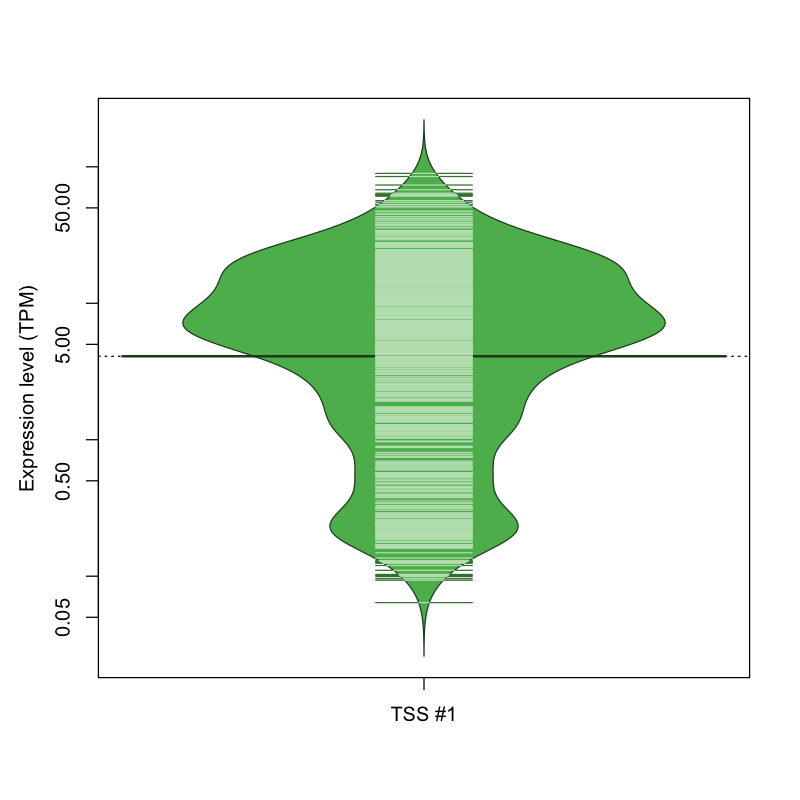
retrocopies.
Species
Parental gene accession
Retrocopies number
Ailuropoda melanoleuca ENSAMEG00000014582 27 retrocopies Show / hide list of retrocopies retro_amel_1008 ,
retro_amel_1182 ,
retro_amel_1185 ,
retro_amel_1218 ,
retro_amel_1277 ,
retro_amel_1342 ,
retro_amel_1419 ,
retro_amel_1549 ,
retro_amel_1638 ,
retro_amel_1642 ,
retro_amel_1753 ,
retro_amel_178 ,
retro_amel_1784 ,
retro_amel_1872 ,
retro_amel_1925 ,
retro_amel_202 ,
retro_amel_220 ,
retro_amel_250 ,
retro_amel_348 ,
retro_amel_52 ,
retro_amel_580 ,
retro_amel_614 ,
retro_amel_624 ,
retro_amel_650 ,
retro_amel_698 ,
retro_amel_861 ,
retro_amel_907 ,
Bos taurus ENSBTAG00000013343 33 retrocopies Show / hide list of retrocopies retro_btau_1007 ,
retro_btau_1011 ,
retro_btau_1033 ,
retro_btau_1197 ,
retro_btau_1209 ,
retro_btau_1277 ,
retro_btau_1354 ,
retro_btau_1396 ,
retro_btau_1452 ,
retro_btau_1464 ,
retro_btau_1466 ,
retro_btau_1481 ,
retro_btau_1511 ,
retro_btau_1540 ,
retro_btau_1561 ,
retro_btau_1683 ,
retro_btau_1781 ,
retro_btau_1798 ,
retro_btau_1848 ,
retro_btau_250 ,
retro_btau_301 ,
retro_btau_339 ,
retro_btau_340 ,
retro_btau_419 ,
retro_btau_422 ,
retro_btau_520 ,
retro_btau_597 ,
retro_btau_639 ,
retro_btau_649 ,
retro_btau_938 ,
retro_btau_963 ,
retro_btau_993 ,
retro_btau_998 ,
Canis familiaris ENSCAFG00000003861 1 retrocopy Show / hide list of retrocopies
Callithrix jacchus ENSCJAG00000000250 13 retrocopies Show / hide list of retrocopies retro_cjac_1171 ,
retro_cjac_1208 ,
retro_cjac_1264 ,
retro_cjac_1643 ,
retro_cjac_2272 ,
retro_cjac_2780 ,
retro_cjac_3113 ,
retro_cjac_3254 ,
retro_cjac_3879 ,
retro_cjac_556 ,
retro_cjac_60 ,
retro_cjac_889 ,
retro_cjac_997 ,
Cavia porcellus ENSCPOG00000020558 1 retrocopy Show / hide list of retrocopies
Dasypus novemcinctus ENSDNOG00000002455 9 retrocopies Show / hide list of retrocopies
Dasypus novemcinctus ENSDNOG00000004126 5 retrocopies Show / hide list of retrocopies
Dasypus novemcinctus ENSDNOG00000005577 36 retrocopies Show / hide list of retrocopies retro_dnov_103 ,
retro_dnov_1164 ,
retro_dnov_1215 ,
retro_dnov_130 ,
retro_dnov_1382 ,
retro_dnov_1441 ,
retro_dnov_1515 ,
retro_dnov_1568 ,
retro_dnov_1657 ,
retro_dnov_1812 ,
retro_dnov_1815 ,
retro_dnov_1918 ,
retro_dnov_1991 ,
retro_dnov_2014 ,
retro_dnov_2134 ,
retro_dnov_2187 ,
retro_dnov_236 ,
retro_dnov_2376 ,
retro_dnov_2379 ,
retro_dnov_247 ,
retro_dnov_2575 ,
retro_dnov_2658 ,
retro_dnov_2667 ,
retro_dnov_285 ,
retro_dnov_305 ,
retro_dnov_432 ,
retro_dnov_439 ,
retro_dnov_543 ,
retro_dnov_598 ,
retro_dnov_642 ,
retro_dnov_807 ,
retro_dnov_841 ,
retro_dnov_846 ,
retro_dnov_849 ,
retro_dnov_861 ,
retro_dnov_897 ,
Dasypus novemcinctus ENSDNOG00000007848 2 retrocopies Show / hide list of retrocopies
Dasypus novemcinctus ENSDNOG00000016683 8 retrocopies Show / hide list of retrocopies
Dipodomys ordii ENSDORG00000015550 5 retrocopies Show / hide list of retrocopies
Felis catus ENSFCAG00000003348 37 retrocopies Show / hide list of retrocopies retro_fcat_101 ,
retro_fcat_1029 ,
retro_fcat_1040 ,
retro_fcat_1059 ,
retro_fcat_1141 ,
retro_fcat_1150 ,
retro_fcat_1295 ,
retro_fcat_1319 ,
retro_fcat_1336 ,
retro_fcat_1359 ,
retro_fcat_139 ,
retro_fcat_1522 ,
retro_fcat_1523 ,
retro_fcat_1602 ,
retro_fcat_1658 ,
retro_fcat_1741 ,
retro_fcat_1747 ,
retro_fcat_1764 ,
retro_fcat_1765 ,
retro_fcat_1766 ,
retro_fcat_1799 ,
retro_fcat_1812 ,
retro_fcat_1901 ,
retro_fcat_297 ,
retro_fcat_298 ,
retro_fcat_315 ,
retro_fcat_343 ,
retro_fcat_50 ,
retro_fcat_533 ,
retro_fcat_536 ,
retro_fcat_620 ,
retro_fcat_675 ,
retro_fcat_68 ,
retro_fcat_778 ,
retro_fcat_918 ,
retro_fcat_927 ,
retro_fcat_964 ,
Homo sapiens ENSG00000087086 11 retrocopies Show / hide list of retrocopies retro_hsap_1647 ,
retro_hsap_1955 ,
retro_hsap_2407 ,
retro_hsap_2484 ,
retro_hsap_2942 ,
retro_hsap_3842 ,
retro_hsap_4146 ,
retro_hsap_431 ,
retro_hsap_4665 ,
retro_hsap_4805 ,
retro_hsap_929 ,
Gorilla gorilla ENSGGOG00000011999 9 retrocopies Show / hide list of retrocopies
Loxodonta africana ENSLAFG00000031248 11 retrocopies Show / hide list of retrocopies retro_lafr_1003 ,
retro_lafr_1064 ,
retro_lafr_1065 ,
retro_lafr_1069 ,
retro_lafr_1206 ,
retro_lafr_255 ,
retro_lafr_345 ,
retro_lafr_346 ,
retro_lafr_785 ,
retro_lafr_836 ,
retro_lafr_977 ,
Microcebus murinus ENSMICG00000001861 7 retrocopies Show / hide list of retrocopies
Macaca mulatta ENSMMUG00000003909 9 retrocopies Show / hide list of retrocopies
Mustela putorius furo ENSMPUG00000003304 29 retrocopies Show / hide list of retrocopies retro_mputfur_1012 ,
retro_mputfur_1026 ,
retro_mputfur_1133 ,
retro_mputfur_1142 ,
retro_mputfur_1150 ,
retro_mputfur_1216 ,
retro_mputfur_1222 ,
retro_mputfur_1225 ,
retro_mputfur_1238 ,
retro_mputfur_1246 ,
retro_mputfur_1268 ,
retro_mputfur_1303 ,
retro_mputfur_1335 ,
retro_mputfur_1521 ,
retro_mputfur_1620 ,
retro_mputfur_195 ,
retro_mputfur_222 ,
retro_mputfur_265 ,
retro_mputfur_350 ,
retro_mputfur_356 ,
retro_mputfur_368 ,
retro_mputfur_482 ,
retro_mputfur_66 ,
retro_mputfur_702 ,
retro_mputfur_741 ,
retro_mputfur_80 ,
retro_mputfur_820 ,
retro_mputfur_906 ,
retro_mputfur_952 ,
Mus musculus ENSMUSG00000024661 4 retrocopies Show / hide list of retrocopies
Mus musculus ENSMUSG00000050708 1 retrocopy Show / hide list of retrocopies retro_mmus_228
Nomascus leucogenys ENSNLEG00000004716 10 retrocopies Show / hide list of retrocopies retro_nleu_1099 ,
retro_nleu_1332 ,
retro_nleu_1871 ,
retro_nleu_1977 ,
retro_nleu_2028 ,
retro_nleu_2180 ,
retro_nleu_2294 ,
retro_nleu_2715 ,
retro_nleu_272 ,
retro_nleu_290 ,
Oryctolagus cuniculus ENSOCUG00000000014 6 retrocopies Show / hide list of retrocopies
Ochotona princeps ENSOPRG00000002492 10 retrocopies Show / hide list of retrocopies retro_opri_187 ,
retro_opri_270 ,
retro_opri_422 ,
retro_opri_43 ,
retro_opri_551 ,
retro_opri_618 ,
retro_opri_702 ,
retro_opri_785 ,
retro_opri_883 ,
retro_opri_886 ,
Procavia capensis ENSPCAG00000013227 6 retrocopies Show / hide list of retrocopies
Pongo abelii ENSPPYG00000010223 11 retrocopies Show / hide list of retrocopies retro_pabe_1362 ,
retro_pabe_1615 ,
retro_pabe_1690 ,
retro_pabe_1814 ,
retro_pabe_2514 ,
retro_pabe_291 ,
retro_pabe_3056 ,
retro_pabe_3463 ,
retro_pabe_3626 ,
retro_pabe_3734 ,
retro_pabe_800 ,
Pan troglodytes ENSPTRG00000011268 11 retrocopies Show / hide list of retrocopies retro_ptro_1118 ,
retro_ptro_1307 ,
retro_ptro_1437 ,
retro_ptro_2055 ,
retro_ptro_2619 ,
retro_ptro_2818 ,
retro_ptro_3101 ,
retro_ptro_3195 ,
retro_ptro_328 ,
retro_ptro_645 ,
retro_ptro_98 ,
Pteropus vampyrus ENSPVAG00000009570 25 retrocopies Show / hide list of retrocopies retro_pvam_1054 ,
retro_pvam_1076 ,
retro_pvam_1205 ,
retro_pvam_1228 ,
retro_pvam_1241 ,
retro_pvam_1270 ,
retro_pvam_1276 ,
retro_pvam_1311 ,
retro_pvam_1383 ,
retro_pvam_1385 ,
retro_pvam_1471 ,
retro_pvam_1479 ,
retro_pvam_229 ,
retro_pvam_29 ,
retro_pvam_314 ,
retro_pvam_322 ,
retro_pvam_377 ,
retro_pvam_41 ,
retro_pvam_447 ,
retro_pvam_47 ,
retro_pvam_491 ,
retro_pvam_519 ,
retro_pvam_542 ,
retro_pvam_677 ,
retro_pvam_959 ,
Rattus norvegicus ENSRNOG00000020843 7 retrocopies Show / hide list of retrocopies
Sorex araneus ENSSARG00000011741 12 retrocopies Show / hide list of retrocopies retro_sara_264 ,
retro_sara_315 ,
retro_sara_359 ,
retro_sara_384 ,
retro_sara_404 ,
retro_sara_408 ,
retro_sara_476 ,
retro_sara_486 ,
retro_sara_506 ,
retro_sara_634 ,
retro_sara_728 ,
retro_sara_87 ,
Sus scrofa ENSSSCG00000003153 33 retrocopies Show / hide list of retrocopies retro_sscr_1040 ,
retro_sscr_1095 ,
retro_sscr_1101 ,
retro_sscr_1142 ,
retro_sscr_1189 ,
retro_sscr_1214 ,
retro_sscr_1215 ,
retro_sscr_124 ,
retro_sscr_197 ,
retro_sscr_212 ,
retro_sscr_224 ,
retro_sscr_305 ,
retro_sscr_331 ,
retro_sscr_400 ,
retro_sscr_531 ,
retro_sscr_532 ,
retro_sscr_533 ,
retro_sscr_534 ,
retro_sscr_535 ,
retro_sscr_536 ,
retro_sscr_559 ,
retro_sscr_589 ,
retro_sscr_605 ,
retro_sscr_74 ,
retro_sscr_747 ,
retro_sscr_799 ,
retro_sscr_817 ,
retro_sscr_830 ,
retro_sscr_833 ,
retro_sscr_839 ,
retro_sscr_840 ,
retro_sscr_862 ,
retro_sscr_898 ,
Ictidomys tridecemlineatus ENSSTOG00000020639 7 retrocopies Show / hide list of retrocopies
Tupaia belangeri ENSTBEG00000005311 13 retrocopies Show / hide list of retrocopies retro_tbel_1144 ,
retro_tbel_1624 ,
retro_tbel_1889 ,
retro_tbel_2263 ,
retro_tbel_263 ,
retro_tbel_2652 ,
retro_tbel_2802 ,
retro_tbel_2916 ,
retro_tbel_3508 ,
retro_tbel_3622 ,
retro_tbel_3735 ,
retro_tbel_795 ,
retro_tbel_931 ,
Tarsius syrichta ENSTSYG00000013988 56 retrocopies Show / hide list of retrocopies retro_tsyr_1035 ,
retro_tsyr_1059 ,
retro_tsyr_106 ,
retro_tsyr_1111 ,
retro_tsyr_1131 ,
retro_tsyr_1139 ,
retro_tsyr_1194 ,
retro_tsyr_1228 ,
retro_tsyr_1241 ,
retro_tsyr_1248 ,
retro_tsyr_1306 ,
retro_tsyr_1315 ,
retro_tsyr_1334 ,
retro_tsyr_1348 ,
retro_tsyr_135 ,
retro_tsyr_1362 ,
retro_tsyr_1372 ,
retro_tsyr_160 ,
retro_tsyr_1716 ,
retro_tsyr_1794 ,
retro_tsyr_1828 ,
retro_tsyr_1841 ,
retro_tsyr_1846 ,
retro_tsyr_1885 ,
retro_tsyr_1975 ,
retro_tsyr_1992 ,
retro_tsyr_2009 ,
retro_tsyr_2046 ,
retro_tsyr_2055 ,
retro_tsyr_228 ,
retro_tsyr_233 ,
retro_tsyr_301 ,
retro_tsyr_341 ,
retro_tsyr_353 ,
retro_tsyr_354 ,
retro_tsyr_43 ,
retro_tsyr_435 ,
retro_tsyr_441 ,
retro_tsyr_489 ,
retro_tsyr_509 ,
retro_tsyr_517 ,
retro_tsyr_618 ,
retro_tsyr_648 ,
retro_tsyr_65 ,
retro_tsyr_661 ,
retro_tsyr_663 ,
retro_tsyr_729 ,
retro_tsyr_758 ,
retro_tsyr_777 ,
retro_tsyr_861 ,
retro_tsyr_925 ,
retro_tsyr_932 ,
retro_tsyr_945 ,
retro_tsyr_947 ,
retro_tsyr_962 ,
retro_tsyr_964 ,
Tursiops truncatus ENSTTRG00000001252 66 retrocopies Show / hide list of retrocopies retro_ttru_1000 ,
retro_ttru_1071 ,
retro_ttru_108 ,
retro_ttru_1092 ,
retro_ttru_1121 ,
retro_ttru_1134 ,
retro_ttru_1191 ,
retro_ttru_1197 ,
retro_ttru_1208 ,
retro_ttru_1214 ,
retro_ttru_1230 ,
retro_ttru_1234 ,
retro_ttru_1243 ,
retro_ttru_1258 ,
retro_ttru_1259 ,
retro_ttru_1260 ,
retro_ttru_1280 ,
retro_ttru_1284 ,
retro_ttru_1313 ,
retro_ttru_1327 ,
retro_ttru_1352 ,
retro_ttru_146 ,
retro_ttru_1460 ,
retro_ttru_1461 ,
retro_ttru_1487 ,
retro_ttru_1489 ,
retro_ttru_1517 ,
retro_ttru_1547 ,
retro_ttru_1574 ,
retro_ttru_1590 ,
retro_ttru_1636 ,
retro_ttru_1642 ,
retro_ttru_1657 ,
retro_ttru_1675 ,
retro_ttru_1688 ,
retro_ttru_1728 ,
retro_ttru_1751 ,
retro_ttru_1753 ,
retro_ttru_1761 ,
retro_ttru_1791 ,
retro_ttru_1805 ,
retro_ttru_1842 ,
retro_ttru_1870 ,
retro_ttru_1936 ,
retro_ttru_201 ,
retro_ttru_320 ,
retro_ttru_321 ,
retro_ttru_343 ,
retro_ttru_346 ,
retro_ttru_39 ,
retro_ttru_452 ,
retro_ttru_525 ,
retro_ttru_665 ,
retro_ttru_743 ,
retro_ttru_753 ,
retro_ttru_759 ,
retro_ttru_797 ,
retro_ttru_811 ,
retro_ttru_823 ,
retro_ttru_835 ,
retro_ttru_847 ,
retro_ttru_877 ,
retro_ttru_895 ,
retro_ttru_939 ,
retro_ttru_943 ,
retro_ttru_960 ,
Vicugna pacos ENSVPAG00000005509 3 retrocopies Show / hide list of retrocopies