RetrogeneDB ID: | retro_mmus_2882 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 6:135349410..135349683(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000082691 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Dynlt1f | ||
| Ensembl ID: | ENSMUSG00000095677 | ||
| Aliases: | Dynlt1f, 100040631, Dynlt1e | ||
| Description: | dynein light chain Tctex-type 1F [Source:MGI Symbol;Acc:MGI:3780996] |

Retrocopy-Parental alignment summary:
>retro_mmus_2882
GAGGCTATAGAAAGCGCCATCGGTGGTAATGCCTACCAGCACAGCAAAGTCAACCAGTGGACCACTAATGTCCTAGAACA
GACTTTGAGCCAACTCACCAAACTGGGGAGACCATTTAAATACATTGTGACCTGTGTGATCATGCAGAAGAACGGTGCTG
GGTTACACTCCGCAAGTTCCTGCTTCTGGGACAGCTCCACAGACGGAAGCTGCACAGTCCGATGGGAGAACAAGACCATG
TACTGCATCGTCAGTACCTTCGGACTGTCCATC
GAGGCTATAGAAAGCGCCATCGGTGGTAATGCCTACCAGCACAGCAAAGTCAACCAGTGGACCACTAATGTCCTAGAACA
GACTTTGAGCCAACTCACCAAACTGGGGAGACCATTTAAATACATTGTGACCTGTGTGATCATGCAGAAGAACGGTGCTG
GGTTACACTCCGCAAGTTCCTGCTTCTGGGACAGCTCCACAGACGGAAGCTGCACAGTCCGATGGGAGAACAAGACCATG
TACTGCATCGTCAGTACCTTCGGACTGTCCATC
ORF - retro_mmus_2882 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 100. % |
| Parental protein coverage: | 80.53 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | EAIESAIGGNAYQHSKVNQWTTNVLEQTLSQLTKLGRPFKYIVTCVIMQKNGAGLHSASSCFWDSSTDGS |
| EAIESAIGGNAYQHSKVNQWTTNVLEQTLSQLTKLGRPFKYIVTCVIMQKNGAGLHSASSCFWDSSTDGS | |
| Retrocopy | EAIESAIGGNAYQHSKVNQWTTNVLEQTLSQLTKLGRPFKYIVTCVIMQKNGAGLHSASSCFWDSSTDGS |
| Parental | CTVRWENKTMYCIVSTFGLSI |
| CTVRWENKTMYCIVSTFGLSI | |
| Retrocopy | CTVRWENKTMYCIVSTFGLSI |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .07 RPM | 0 .21 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 0 .48 RPM |
| SRP007412_heart | 0 .12 RPM | 0 .28 RPM |
| SRP007412_kidney | 0 .12 RPM | 0 .48 RPM |
| SRP007412_liver | 0 .00 RPM | 0 .16 RPM |
| SRP007412_testis | 1 .28 RPM | 12 .35 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_2882 was not detected
No EST(s) were mapped for retro_mmus_2882 retrocopy.
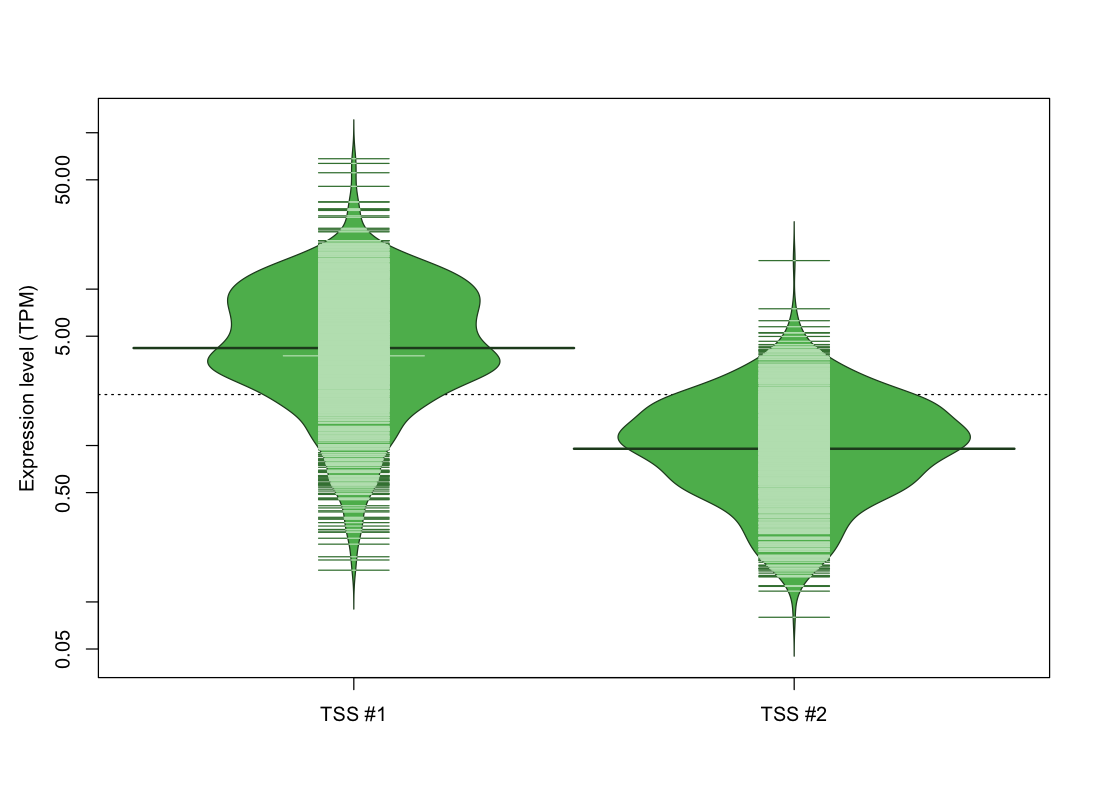
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_119624 | 104 libraries | 79 libraries | 452 libraries | 254 libraries | 183 libraries |
| TSS #2 | TSS_119625 | 241 libraries | 405 libraries | 420 libraries | 5 libraries | 1 library |
The graphical summary, for retro_mmus_2882 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2882 was not experimentally validated.