RetrogeneDB ID: | retro_hsap_2674 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 3:96068397..96069787(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000243547 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | HNRNPK | ||
| Ensembl ID: | ENSG00000165119 | ||
| Aliases: | HNRNPK, CSBP, HNRPK, TUNP | ||
| Description: | heterogeneous nuclear ribonucleoprotein K [Source:HGNC Symbol;Acc:5044] |

Retrocopy-Parental alignment summary:
>retro_hsap_2674
ATGTAAACTGAACAGCCGGAAGAAACCTTCCCGAACACTGAAACCAATGGTGAATTTGGTAAACACCCTGCAGAAGATAT
GGAAGAGTAACAAGCATTTAAAAGATCTAGAAACACTGATGAGATGGTTGAATTACGCATTCTGCTTCAGACCAAGAATG
CTGGGGCAGTGATTGGAAAAGGAGGCAAGAATATCAAGGCTCTCCGTACAGACTACAATGCCAGTGTTTCAGTCCCAGAC
AGCAGTGGCCCCGAGCACATATTGAGTATCAGTGCTGGTATTGACACAATTGGAGAAATTCTGAAGAAAATCATCCCTAC
CTTGGAAGAGGGCCTGCAGTTTCCATCACCCACTGCAACCAGCCAGCTGCCGCTCGAATCTGTTGCTGTGGAATGCTTAA
GTTACCAACACTATAAAGGAAGTGACTTTGACTGCGAGTTGAGGCTGTTGATTCATCAGAGTCTAGCAGGAGGAATTATT
GGGGTCGAAGGTGCTAAAATCAAAGAACTTCGAGAGAACACTCAAACCACCATCAAGCTTTTCCAGGAATGCTGTCCTTA
TTCCACTGACAAGAGTTGTTCTTATTGGAGGAAAACCCGATAGGGTTGTAGAGTGCATAAAGATCATCCTTGATCTTATA
TATGAGTCTCCCATCAAAGGACGTGCACAGCCTTATGATCCCAATTTTTATGATGAAACCTATGATTATGGTGGTTTTAC
AATTATGTTTGATGACCGCTGTGGACGCCCAGTGGGATTTCCCATGCGGGGAAGAGGTGGTTTTGACAGAATGCCTCCTG
GTCAGGGTGGGCGTCCCATGCCTCCATCTAGAAGAGATTATGATGATATGAGCCCTCGTCAAGGACCACCTCCACCTCCT
CCAGGACAAGGTGGTGAGGGTGGTAGCAGAGCTCGGAATCTTCCTCTTCCTCCTCCACCACCACCTAGAGGGGGAGACCT
CATGGCCTATGACAGAAGAGGGAGACCTGGAGACTGTTACCATGGCATGGTTGGTTTCAGTGCTGATGAAACTTGGGACT
CTGCAATAGATACATGGAGCCCATCAGAATGGCAGATGGCTTATGAACCACAGGGTGGCTCCAGATATGATTATTCCTAT
GCAGGGGGTCGTGGCTCATATGGTAATCTTGGTGGACTTATTATTACTACACAAGTAACGATTCCCAAAGATTTGGCTGG
ATCTATTATTGGCAAAGGTGGTCTGCAGATTAAACAAATCCGTCATGAGTCGGGAGCTTCGATCAAAATTGATGAGCCTT
TAGAAGGATTGGAAGATTGGATCATTACCATTAGAGAAACACAGGACCAGATACAGAATGCACAGTATTTGCTGCAGAAC
AGTGTGAAGCAGTGTGCAGATGTTGAAGGA
ATGTAAACTGAACAGCCGGAAGAAACCTTCCCGAACACTGAAACCAATGGTGAATTTGGTAAACACCCTGCAGAAGATAT
GGAAGAGTAACAAGCATTTAAAAGATCTAGAAACACTGATGAGATGGTTGAATTACGCATTCTGCTTCAGACCAAGAATG
CTGGGGCAGTGATTGGAAAAGGAGGCAAGAATATCAAGGCTCTCCGTACAGACTACAATGCCAGTGTTTCAGTCCCAGAC
AGCAGTGGCCCCGAGCACATATTGAGTATCAGTGCTGGTATTGACACAATTGGAGAAATTCTGAAGAAAATCATCCCTAC
CTTGGAAGAGGGCCTGCAGTTTCCATCACCCACTGCAACCAGCCAGCTGCCGCTCGAATCTGTTGCTGTGGAATGCTTAA
GTTACCAACACTATAAAGGAAGTGACTTTGACTGCGAGTTGAGGCTGTTGATTCATCAGAGTCTAGCAGGAGGAATTATT
GGGGTCGAAGGTGCTAAAATCAAAGAACTTCGAGAGAACACTCAAACCACCATCAAGCTTTTCCAGGAATGCTGTCCTTA
TTCCACTGACAAGAGTTGTTCTTATTGGAGGAAAACCCGATAGGGTTGTAGAGTGCATAAAGATCATCCTTGATCTTATA
TATGAGTCTCCCATCAAAGGACGTGCACAGCCTTATGATCCCAATTTTTATGATGAAACCTATGATTATGGTGGTTTTAC
AATTATGTTTGATGACCGCTGTGGACGCCCAGTGGGATTTCCCATGCGGGGAAGAGGTGGTTTTGACAGAATGCCTCCTG
GTCAGGGTGGGCGTCCCATGCCTCCATCTAGAAGAGATTATGATGATATGAGCCCTCGTCAAGGACCACCTCCACCTCCT
CCAGGACAAGGTGGTGAGGGTGGTAGCAGAGCTCGGAATCTTCCTCTTCCTCCTCCACCACCACCTAGAGGGGGAGACCT
CATGGCCTATGACAGAAGAGGGAGACCTGGAGACTGTTACCATGGCATGGTTGGTTTCAGTGCTGATGAAACTTGGGACT
CTGCAATAGATACATGGAGCCCATCAGAATGGCAGATGGCTTATGAACCACAGGGTGGCTCCAGATATGATTATTCCTAT
GCAGGGGGTCGTGGCTCATATGGTAATCTTGGTGGACTTATTATTACTACACAAGTAACGATTCCCAAAGATTTGGCTGG
ATCTATTATTGGCAAAGGTGGTCTGCAGATTAAACAAATCCGTCATGAGTCGGGAGCTTCGATCAAAATTGATGAGCCTT
TAGAAGGATTGGAAGATTGGATCATTACCATTAGAGAAACACAGGACCAGATACAGAATGCACAGTATTTGCTGCAGAAC
AGTGTGAAGCAGTGTGCAGATGTTGAAGGA
ORF - retro_hsap_2674 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.1 % |
| Parental protein coverage: | 99.78 % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | METEQPEETFPNTETNGEFGKRPAEDMEEEQAFKRSRNTDEMVELRILLQSKNAGAVIGKGGKNIKALRT |
| M.TEQPEETFPNTETNGEFGK.PAEDMEE.QAFKRSRNTDEMVELRILLQ.KNAGAVIGKGGKNIKALRT | |
| Retrocopy | M*TEQPEETFPNTETNGEFGKHPAEDMEE*QAFKRSRNTDEMVELRILLQTKNAGAVIGKGGKNIKALRT |
| Parental | DYNASVSVPDSSGPERILSISADIETIGEILKKIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQHYKG |
| DYNASVSVPDSSGPE.ILSISA.I.TIGEILKKIIPTLEEGLQ.PSPTATSQLPLES.AVECL.YQHYKG | |
| Retrocopy | DYNASVSVPDSSGPEHILSISAGIDTIGEILKKIIPTLEEGLQFPSPTATSQLPLESVAVECLSYQHYKG |
| Parental | SDFDCELRLLIHQSLAGGIIGVKGAKIKELRENTQTTIKLFQECCPHSTD-RVVLIGGKPDRVVECIKII |
| SDFDCELRLLIHQSLAGGIIGV.GAKIKELRENTQTTIKLFQECCP.STD.RVVLIGGKPDRVVECIKII | |
| Retrocopy | SDFDCELRLLIHQSLAGGIIGVEGAKIKELRENTQTTIKLFQECCPYSTD>RVVLIGGKPDRVVECIKII |
| Parental | LDLISESPIKGRAQPYDPNFYDETYDYGGFTMMFDDRRGRPVGFPMRGRGGFDRMPPGRGGRPMPPSRRD |
| LDLI.ESPIKGRAQPYDPNFYDETYDYGGFT.MFDDR.GRPVGFPMRGRGGFDRMPPG.GGRPMPPSRRD | |
| Retrocopy | LDLIYESPIKGRAQPYDPNFYDETYDYGGFTIMFDDRCGRPVGFPMRGRGGFDRMPPGQGGRPMPPSRRD |
| Parental | YDDMSPRRGPPPPPPGRGGRGGSRARNLPLPPPPPPRGGDLMAYDRRGRPGDRYDGMVGFSADETWDSAI |
| YDDMSPR.GPPPPPPG.GG.GGSRARNLPLPPPPPPRGGDLMAYDRRGRPGD.Y.GMVGFSADETWDSAI | |
| Retrocopy | YDDMSPRQGPPPPPPGQGGEGGSRARNLPLPPPPPPRGGDLMAYDRRGRPGDCYHGMVGFSADETWDSAI |
| Parental | DTWSPSEWQMAYEPQGGSGYDYSYAGGRGSYGDLGGPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGA |
| DTWSPSEWQMAYEPQGGS.YDYSYAGGRGSYG.LGG.IITTQVTIPKDLAGSIIGKGG..IKQIRHESGA | |
| Retrocopy | DTWSPSEWQMAYEPQGGSRYDYSYAGGRGSYGNLGGLIITTQVTIPKDLAGSIIGKGGLQIKQIRHESGA |
| Parental | SIKIDEPLEGSEDRIITITGTQDQIQNAQYLLQNSVKQYADVEG |
| SIKIDEPLEG.ED.IITI..TQDQIQNAQYLLQNSVKQ.ADVEG | |
| Retrocopy | SIKIDEPLEGLEDWIITIRETQDQIQNAQYLLQNSVKQCADVEG |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .26 RPM | 632 .24 RPM |
| bodymap2_adrenal | 0 .25 RPM | 629 .54 RPM |
| bodymap2_brain | 0 .35 RPM | 489 .31 RPM |
| bodymap2_breast | 0 .10 RPM | 490 .86 RPM |
| bodymap2_colon | 0 .02 RPM | 535 .89 RPM |
| bodymap2_heart | 0 .09 RPM | 269 .31 RPM |
| bodymap2_kidney | 0 .23 RPM | 464 .01 RPM |
| bodymap2_liver | 0 .06 RPM | 257 .46 RPM |
| bodymap2_lung | 0 .05 RPM | 592 .28 RPM |
| bodymap2_lymph_node | 0 .16 RPM | 446 .35 RPM |
| bodymap2_ovary | 0 .23 RPM | 648 .99 RPM |
| bodymap2_prostate | 0 .21 RPM | 490 .85 RPM |
| bodymap2_skeletal_muscle | 0 .22 RPM | 283 .35 RPM |
| bodymap2_testis | 0 .13 RPM | 574 .25 RPM |
| bodymap2_thyroid | 0 .32 RPM | 702 .41 RPM |
| bodymap2_white_blood_cells | 0 .16 RPM | 806 .90 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_2674 was not detected
No EST(s) were mapped for retro_hsap_2674 retrocopy.
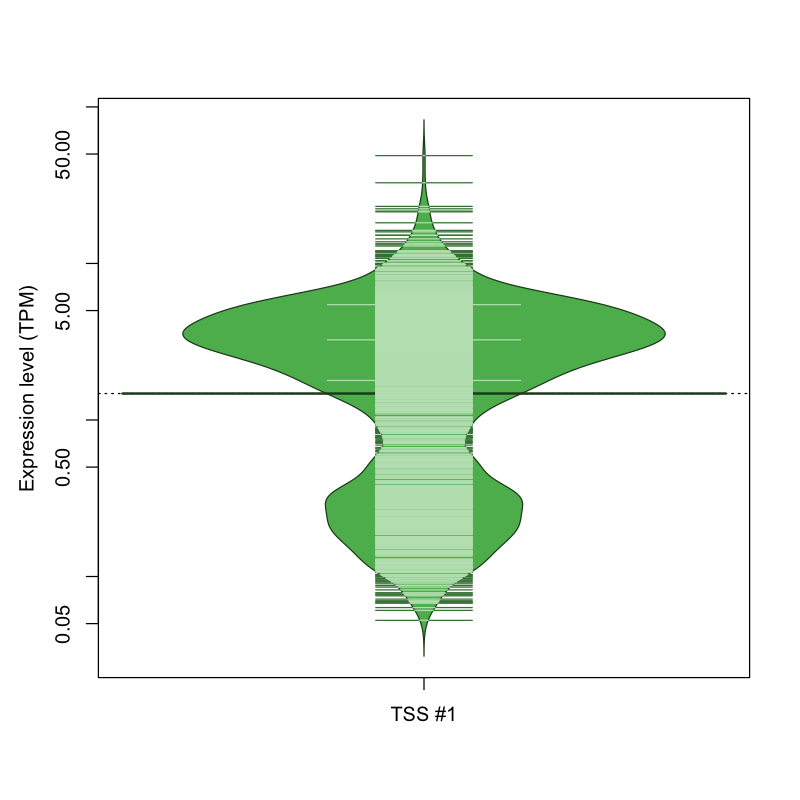
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_137861 | 629 libraries | 397 libraries | 605 libraries | 167 libraries | 31 libraries |
The graphical summary, for retro_hsap_2674 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_2674 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2674 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pongo abelii | retro_pabe_42 |
Parental genes homology:
Parental genes homology involve 14 parental genes, and 32 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000021131 | 2 retrocopies | |
| Homo sapiens | ENSG00000165119 | 3 retrocopies |
retro_hsap_2323, retro_hsap_2674 , retro_hsap_3385,
|
| Homo sapiens | ENSG00000197111 | 2 retrocopies | |
| Microcebus murinus | ENSMICG00000015780 | 3 retrocopies | |
| Myotis lucifugus | ENSMLUG00000005871 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000018871 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000008402 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000021546 | 2 retrocopies | |
| Otolemur garnettii | ENSOGAG00000007478 | 3 retrocopies | |
| Pongo abelii | ENSPPYG00000019327 | 3 retrocopies | |
| Sarcophilus harrisii | ENSSHAG00000017109 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000001289 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000004906 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000016693 | 7 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .94 RPM |
| CEU_NA11843 | 0 .54 RPM |
| CEU_NA11930 | 1 .34 RPM |
| CEU_NA12004 | 0 .57 RPM |
| CEU_NA12400 | 0 .60 RPM |
| CEU_NA12751 | 0 .44 RPM |
| CEU_NA12760 | 1 .17 RPM |
| CEU_NA12827 | 0 .52 RPM |
| CEU_NA12872 | 0 .38 RPM |
| CEU_NA12873 | 0 .96 RPM |
| FIN_HG00183 | 1 .15 RPM |
| FIN_HG00277 | 1 .04 RPM |
| FIN_HG00315 | 0 .47 RPM |
| FIN_HG00321 | 0 .70 RPM |
| FIN_HG00328 | 0 .45 RPM |
| FIN_HG00338 | 0 .64 RPM |
| FIN_HG00349 | 1 .28 RPM |
| FIN_HG00375 | 0 .81 RPM |
| FIN_HG00377 | 0 .81 RPM |
| FIN_HG00378 | 0 .66 RPM |
| GBR_HG00099 | 0 .93 RPM |
| GBR_HG00111 | 0 .58 RPM |
| GBR_HG00114 | 0 .47 RPM |
| GBR_HG00119 | 0 .24 RPM |
| GBR_HG00131 | 0 .86 RPM |
| GBR_HG00133 | 0 .51 RPM |
| GBR_HG00134 | 0 .59 RPM |
| GBR_HG00137 | 0 .66 RPM |
| GBR_HG00142 | 0 .61 RPM |
| GBR_HG00143 | 0 .32 RPM |
| TSI_NA20512 | 0 .51 RPM |
| TSI_NA20513 | 0 .65 RPM |
| TSI_NA20518 | 1 .55 RPM |
| TSI_NA20532 | 0 .99 RPM |
| TSI_NA20538 | 0 .51 RPM |
| TSI_NA20756 | 1 .37 RPM |
| TSI_NA20765 | 0 .56 RPM |
| TSI_NA20771 | 1 .25 RPM |
| TSI_NA20786 | 0 .81 RPM |
| TSI_NA20798 | 0 .59 RPM |
| YRI_NA18870 | 1 .05 RPM |
| YRI_NA18907 | 0 .73 RPM |
| YRI_NA18916 | 0 .48 RPM |
| YRI_NA19093 | 0 .57 RPM |
| YRI_NA19099 | 1 .09 RPM |
| YRI_NA19114 | 0 .66 RPM |
| YRI_NA19118 | 0 .62 RPM |
| YRI_NA19213 | 0 .65 RPM |
| YRI_NA19214 | 0 .59 RPM |
| YRI_NA19223 | 0 .86 RPM |