RetrogeneDB ID: | retro_mmus_16 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:95025444..95026194(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000071722 | |
| Aliases: | Spin4, 9630042H07Rik | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Spin1 | ||
| Ensembl ID: | ENSMUSG00000021395 | ||
| Aliases: | Spin1, Spin | ||
| Description: | spindlin 1 [Source:MGI Symbol;Acc:MGI:109242] |

Retrocopy-Parental alignment summary:
>retro_mmus_16
ATGTCTCCTCCGACTGTGCCTCCTACTGGGGTGGATGGTGTGTCCGCGTACCTGATGAAGAAAAGGCACACTCACAAGAA
GCAGCGCCGTAAGCCCACTTTCCTCACTCATAGGAACATCGTGGGCTGCCGCATTCAACACGGCTGGAAAGAAGGCAATG
AGCCCGTGGAGCAATGGAAAGGTACTGTGCTCGAGCAGGTCTCCGTGAAGCCCACTCTGTATATCATCAAATATGATGGC
AAAGACAGTGTGTACGGACTGGAACTGCACAGAGATAAGAGAGTTTTAGCGCTGGAGATTCTTCCTGAGAGAGTTCCTTC
TCCTCGCATTGACTCTCGCCTGGCCGATTCCCTGGTTGGGAAGGCAGTGGGACATGTGTTCGAAGGTGAGCATGGCAAGA
AAGATGAGTGGAAGGGTATGGTGTTGGCGCGAGCCCCCATCATGGATACTTGGTTTTACATCACCTACGAGAAAGATCCG
GTCCTCTATATGTACACCCTGCTGGACGACTACAAAGATGGTGACCTGCGTATCATTCCAGATTCCAACTACTACTTCCC
TGCAGCTGAACAGGAGCCTGGGGAGGTGCTCGACAGCCTTGTGGGCAAGCAGGTGGAACACGCCAAAGACGACGGGTCCA
AGAGAACTGGGATCTTTATTCATCAGGTGGTAGCCAAGCCATCTGTCTACTTCATTAAGTTTGATGATGATATTCACATT
TATGTCTATGGCTTGGTGAAAACCCCGTAA
ATGTCTCCTCCGACTGTGCCTCCTACTGGGGTGGATGGTGTGTCCGCGTACCTGATGAAGAAAAGGCACACTCACAAGAA
GCAGCGCCGTAAGCCCACTTTCCTCACTCATAGGAACATCGTGGGCTGCCGCATTCAACACGGCTGGAAAGAAGGCAATG
AGCCCGTGGAGCAATGGAAAGGTACTGTGCTCGAGCAGGTCTCCGTGAAGCCCACTCTGTATATCATCAAATATGATGGC
AAAGACAGTGTGTACGGACTGGAACTGCACAGAGATAAGAGAGTTTTAGCGCTGGAGATTCTTCCTGAGAGAGTTCCTTC
TCCTCGCATTGACTCTCGCCTGGCCGATTCCCTGGTTGGGAAGGCAGTGGGACATGTGTTCGAAGGTGAGCATGGCAAGA
AAGATGAGTGGAAGGGTATGGTGTTGGCGCGAGCCCCCATCATGGATACTTGGTTTTACATCACCTACGAGAAAGATCCG
GTCCTCTATATGTACACCCTGCTGGACGACTACAAAGATGGTGACCTGCGTATCATTCCAGATTCCAACTACTACTTCCC
TGCAGCTGAACAGGAGCCTGGGGAGGTGCTCGACAGCCTTGTGGGCAAGCAGGTGGAACACGCCAAAGACGACGGGTCCA
AGAGAACTGGGATCTTTATTCATCAGGTGGTAGCCAAGCCATCTGTCTACTTCATTAAGTTTGATGATGATATTCACATT
TATGTCTATGGCTTGGTGAAAACCCCGTAA
ORF - retro_mmus_16 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 72.93 % |
| Parental protein coverage: | 87.02 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | KKHRTSVGPSKPVSQPRRNIVGCRIQHGWREGNGPVTQWKGTVLDQVPVNPSLYLIKYDGFDCVYGLELN |
| K.H.......KP.....RNIVGCRIQHGW.EGN.PV.QWKGTVL.QV.V.P.LY.IKYDG.D.VYGLEL. | |
| Retrocopy | KRHTHKKQRRKPTFLTHRNIVGCRIQHGWKEGNEPVEQWKGTVLEQVSVKPTLYIIKYDGKDSVYGLELH |
| Parental | KDERVSALEVLPDRVATSRISDAHLADTMIGKAVEHMFETEDGSKDEWRGMVLARAPVMNTWFYITYEKD |
| .D.RV.ALE.LP.RV...RI.D..LAD...GKAV.H.FE.E.G.KDEW.GMVLARAP.M.TWFYITYEKD | |
| Retrocopy | RDKRVLALEILPERVPSPRI-DSRLADSLVGKAVGHVFEGEHGKKDEWKGMVLARAPIMDTWFYITYEKD |
| Parental | PVLYMYQLLDDYKEGDLRIMPDSN-DSPPAEREPGEVVDSLVGKQVEYAKEDGSKRTGMVIHQVEAKPSV |
| PVLYMY.LLDDYK.GDLRI.PDSN...P.AE.EPGEV.DSLVGKQVE.AK.DGSKRTG..IHQV.AKPSV | |
| Retrocopy | PVLYMYTLLDDYKDGDLRIIPDSNYYFPAAEQEPGEVLDSLVGKQVEHAKDDGSKRTGIFIHQVVAKPSV |
| Parental | YFIKFDDDFHIYVYDLVKT |
| YFIKFDDD.HIYVY.LVKT | |
| Retrocopy | YFIKFDDDIHIYVYGLVKT |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .14 RPM | 167 .51 RPM |
| SRP007412_cerebellum | 0 .43 RPM | 95 .61 RPM |
| SRP007412_heart | 0 .53 RPM | 31 .35 RPM |
| SRP007412_kidney | 0 .48 RPM | 70 .16 RPM |
| SRP007412_liver | 0 .14 RPM | 54 .51 RPM |
| SRP007412_testis | 0 .09 RPM | 23 .42 RPM |
RNA Polymerase II actvity may be related with retro_mmus_16 in 2 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | X:95026408..95026966 |
| ENCFF001YIJ | POLR2A | X:95026168..95027052 |
No EST(s) were mapped for retro_mmus_16 retrocopy.
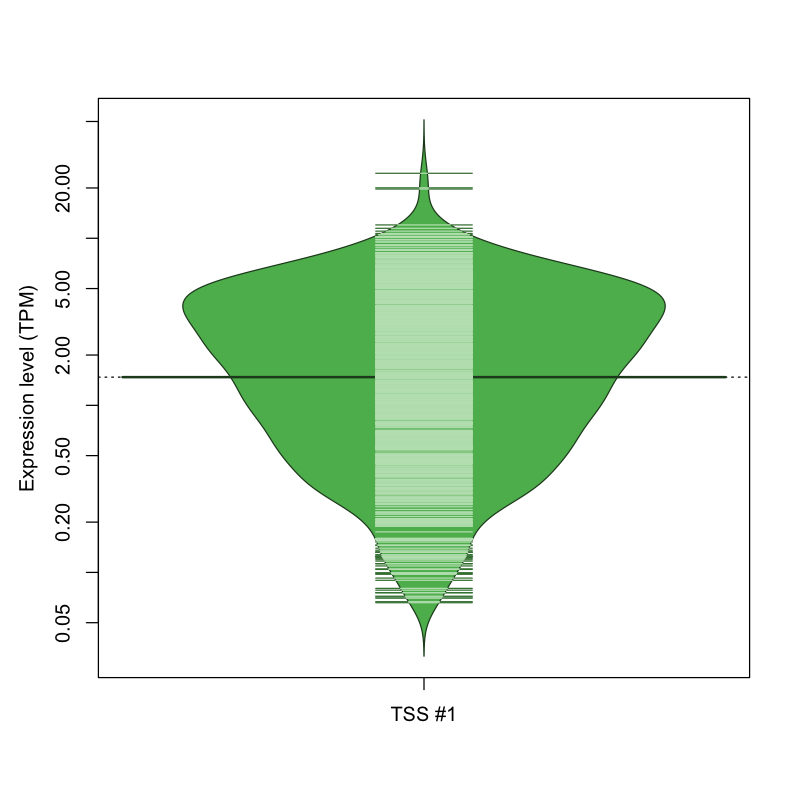
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_158571 | 239 libraries | 294 libraries | 410 libraries | 117 libraries | 12 libraries |
The graphical summary, for retro_mmus_16 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_16 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_16 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_281 |