RetrogeneDB ID: | retro_mmus_190 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:157567631..157568768(-) | ||
| Located in intron of: | ENSMUSG00000046873 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000091736 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Yy1 | ||
| Ensembl ID: | ENSMUSG00000021264 | ||
| Aliases: | Yy1, AW488674, NF-E1 | ||
| Description: | YY1 transcription factor [Source:MGI Symbol;Acc:MGI:99150] |

Retrocopy-Parental alignment summary:
>retro_mmus_190
ATGGCCTCTGAGACAGAGAAACTTCTGTGTCTTAACACAGAAAGCGCAGAGATTCCTGCTGATTTTGTGGAGCTGCTGCC
ACCGGATAATATCGGTGATATCGAGGCGGTTTCTCTGGAAACCAGCGTAGGCCAAACCATCGAAGTATATGGAGATGTAG
GTGTCGATTGGGCGCATGGTAGCCAATACCACTCTCCTGTTATTGCTCTGCAGCCGCTTGTGGGCAGCAGCCTGAGTAGT
AGAGATCATGATAAGGAAATGTTTGTCGTTCAGACACGAGAGGAGGAGGTGGTGGGCTACCAAGACTCAGACAACCTACT
ATTCAGTCCTGAATTTGGAAGCCAGATGGTTTTGCCAGTTAATGAAGACGACTATCTCCAGCCGACCACTGCCTCTTTTA
CGGGCTTTCTGGCGGCAGAGAATGGTCAAGGTGAGCTCAGCCCCTATGAAGGGAATCTCTGTGGTTTGACGACCTTTATA
GAGGCCGGTGCAGAAGAAAGCGTGAACGCTGACCTGGGTGACAAACAGTGGGAGCAGAAGCAGATCGATGGTCTTGATGG
CGAATTTCCCTTCACCATGTGGGACGATGTTAATGAAAAAGAAGATCCCATAGCTGAAGAACAGGCTGGTGAGTCACCCC
CCGATTATTCTGAGTATATGACAGGAAAGAAATTTCCTCCTGAAGGAATACCTGGCATTGATCTTTCTGATCCCAAACAA
CTGGCAGAATTTACTAGCATGAGACCCAAAAAGCCAAAAGGAGACTTTCCAAGACCTATAGCATGCTCTCATAAAGGCTG
CGAGAAGATGTTCAAAGATAATTCTGCTATGAGAAAGCACTTGCACATCCATGGGCCGAGAGTGCATGTCTGTGCAGAAT
GTGGCAAAGCTTTTGTTGAGAGTTCCAAGCTGAAACGTCATCAGCTTGTTCACACTGGAGAGAAACCCTACCAGTGTACC
TTTGAAGGCTGTGGGAGACGCTTTTCCCTGGATTTCAATTTGCGCACACATGTGCGAATCCATACTGGAGACAAACCTTT
CGTGTGTCCATTTGATGCCTGCAACAAGAAGTTTGCACAGTCAACAAACCTGAAATCGCATATCTTAACCCATGTTAAGA
ACAAGAATGACCAGTAA
ATGGCCTCTGAGACAGAGAAACTTCTGTGTCTTAACACAGAAAGCGCAGAGATTCCTGCTGATTTTGTGGAGCTGCTGCC
ACCGGATAATATCGGTGATATCGAGGCGGTTTCTCTGGAAACCAGCGTAGGCCAAACCATCGAAGTATATGGAGATGTAG
GTGTCGATTGGGCGCATGGTAGCCAATACCACTCTCCTGTTATTGCTCTGCAGCCGCTTGTGGGCAGCAGCCTGAGTAGT
AGAGATCATGATAAGGAAATGTTTGTCGTTCAGACACGAGAGGAGGAGGTGGTGGGCTACCAAGACTCAGACAACCTACT
ATTCAGTCCTGAATTTGGAAGCCAGATGGTTTTGCCAGTTAATGAAGACGACTATCTCCAGCCGACCACTGCCTCTTTTA
CGGGCTTTCTGGCGGCAGAGAATGGTCAAGGTGAGCTCAGCCCCTATGAAGGGAATCTCTGTGGTTTGACGACCTTTATA
GAGGCCGGTGCAGAAGAAAGCGTGAACGCTGACCTGGGTGACAAACAGTGGGAGCAGAAGCAGATCGATGGTCTTGATGG
CGAATTTCCCTTCACCATGTGGGACGATGTTAATGAAAAAGAAGATCCCATAGCTGAAGAACAGGCTGGTGAGTCACCCC
CCGATTATTCTGAGTATATGACAGGAAAGAAATTTCCTCCTGAAGGAATACCTGGCATTGATCTTTCTGATCCCAAACAA
CTGGCAGAATTTACTAGCATGAGACCCAAAAAGCCAAAAGGAGACTTTCCAAGACCTATAGCATGCTCTCATAAAGGCTG
CGAGAAGATGTTCAAAGATAATTCTGCTATGAGAAAGCACTTGCACATCCATGGGCCGAGAGTGCATGTCTGTGCAGAAT
GTGGCAAAGCTTTTGTTGAGAGTTCCAAGCTGAAACGTCATCAGCTTGTTCACACTGGAGAGAAACCCTACCAGTGTACC
TTTGAAGGCTGTGGGAGACGCTTTTCCCTGGATTTCAATTTGCGCACACATGTGCGAATCCATACTGGAGACAAACCTTT
CGTGTGTCCATTTGATGCCTGCAACAAGAAGTTTGCACAGTCAACAAACCTGAAATCGCATATCTTAACCCATGTTAAGA
ACAAGAATGACCAGTAA
ORF - retro_mmus_190 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 70. % |
| Parental protein coverage: | 60.39 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | ASSGGGRVKKGGGKKSGKKSYLGGGAGAAGGGGADPGNKKWEQKQVQIKTLEGEFSVTMWSSDEKKDIDH |
| A..G.G......G...G........AGA.....AD.G.K.WEQKQ.....L.GEF..TMW.....K.... | |
| Retrocopy | AENGQGELSPYEGNLCGLTTFIE--AGAEESVNADLGDKQWEQKQID--GLDGEFPFTMWDDVNEKE--- |
| Parental | ETVVEEQIIGENSPPDYSEYMTGKKLPPGGIPGIDLSDPKQLAEFARMKPRKIKEDDAPRTIACPHKGCT |
| ....EEQ..GE.SPPDYSEYMTGKK.PP.GIPGIDLSDPKQLAEF..M.P.K.K..D.PR.IAC.HKGC. | |
| Retrocopy | DPIAEEQA-GE-SPPDYSEYMTGKKFPPEGIPGIDLSDPKQLAEFTSMRPKKPK-GDFPRPIACSHKGCE |
| Parental | KMFRDNSAMRKHLHTHGPRVHVCAECGKAFVESSKLKRHQLVHTGEKPFQCTFEGCGKRFSLDFNLRTHV |
| KMF.DNSAMRKHLH.HGPRVHVCAECGKAFVESSKLKRHQLVHTGEKP.QCTFEGCG.RFSLDFNLRTHV | |
| Retrocopy | KMFKDNSAMRKHLHIHGPRVHVCAECGKAFVESSKLKRHQLVHTGEKPYQCTFEGCGRRFSLDFNLRTHV |
| Parental | RIHTGDRPYVCPFDGCNKKFAQSTNLKSHILTHAKAKNNQ |
| RIHTGD.P.VCPFD.CNKKFAQSTNLKSHILTH.K.KN.Q | |
| Retrocopy | RIHTGDKPFVCPFDACNKKFAQSTNLKSHILTHVKNKNDQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .75 RPM | 26 .98 RPM |
| SRP007412_cerebellum | 1 .04 RPM | 24 .97 RPM |
| SRP007412_heart | 1 .43 RPM | 21 .67 RPM |
| SRP007412_kidney | 1 .39 RPM | 33 .13 RPM |
| SRP007412_liver | 0 .49 RPM | 29 .55 RPM |
| SRP007412_testis | 1 .10 RPM | 30 .78 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_190 was not detected
No EST(s) were mapped for retro_mmus_190 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_156010 | 1007 libraries | 23 libraries | 26 libraries | 13 libraries | 3 libraries |
The graphical summary, for retro_mmus_190 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
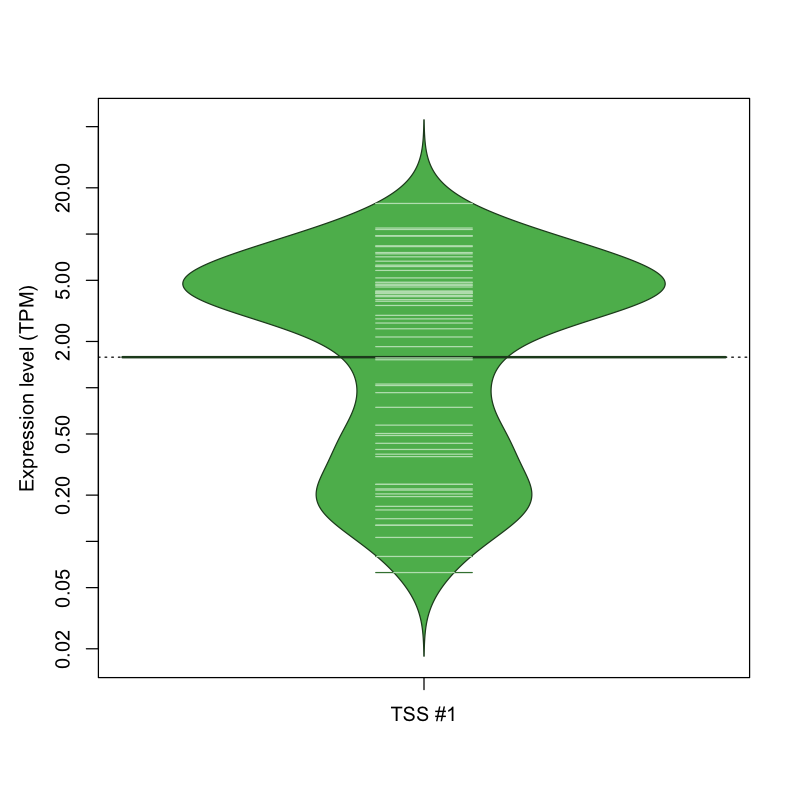
retro_mmus_190 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_190 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_2 |
Parental genes homology:
Parental genes homology involve 14 parental genes, and 27 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000011146 | 1 retrocopy | |
| Bos taurus | ENSBTAG00000020819 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000016853 | 2 retrocopies | |
| Cavia porcellus | ENSCPOG00000020809 | 2 retrocopies | |
| Equus caballus | ENSECAG00000005805 | 1 retrocopy | |
| Homo sapiens | ENSG00000100811 | 2 retrocopies | |
| Gorilla gorilla | ENSGGOG00000026332 | 2 retrocopies | |
| Mustela putorius furo | ENSMPUG00000002931 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000021264 | 1 retrocopy |
retro_mmus_190 ,
|
| Nomascus leucogenys | ENSNLEG00000000999 | 3 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000026938 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000006955 | 4 retrocopies | |
| Pongo abelii | ENSPPYG00000025949 | 3 retrocopies | |
| Pan troglodytes | ENSPTRG00000006717 | 3 retrocopies |