RetrogeneDB ID: | retro_mmus_1052 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 13:98613500..98614491(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Cnn3 | ||
| Ensembl ID: | ENSMUSG00000053931 | ||
| Aliases: | Cnn3, 1600014M03Rik, C85854, Calpo3 | ||
| Description: | calponin 3, acidic [Source:MGI Symbol;Acc:MGI:1919244] |

Retrocopy-Parental alignment summary:
>retro_mmus_1052
ATGACCCACTTCAACAAGGGCCCTTCCTACGGGCTCTCCGCCGAAGTTAAAGAACAAAATCGCATCCAAGTATGACCAGC
AGGCCGAGGAAGATCTGCCTAACTGGATAGAAGAGGTGACAGGCCTAGGCATTGGCACCAACTACCAGCTGGGGCTGAAG
GACGGCATCATCCTGTGCGAACTCATAAACAAGCTACAGTCAGGCTCTGTGAAAAAAGTCAATGAATCCTCACTAAATTG
GCCGCAGTTGGAGAATATCGGCAACTTCATTAAAGCTATCCAGGCTTACGGTATGAAGCCCCATGATATATTTGAAGCAA
ACGAACTCTTTGAGAATGGCAACATGACCCAGGTTCAGACGACACTGGTGGCTCTAGCAGGTCTGGTGAAAACAAAAGGA
TTCCATACAACCATTGACACTGGTGTTAAGTATGCAGAAAAACAAACAAGACGTTTTGATGAAGGCAAATTAAAGGCTGG
CCAGAGTGTAATTGGTTTACAGATGGGTACCAACAAATGTGCCAGCCAAGCGGGCACGACAGCCTATGGGACTCGGAGGC
ATCTTTATGATCCCAAGATGCAGACGGACAAACCCTTTGACCAGACCACGATTAGCCTGCAGATGGGCACCAACAAAGGA
GCCAGCCAGGCTGGGATGTTAACACCAGGTACCAGAAGAGACATCTATGACCAGAAGTTGACATTACAGCCAGTGGACAA
CTCGACCATTTCTCTACAGATGGGCACCAACAAAACTGCTTCCCAGAAAGGAATGAGCGTGTATTGGCTTGGGCGGCAAG
TGTATGACCCCAAGTACTGTGCCGCACCCACAGAACCTGTCATTCACAACGGAAGCCAGGGCACGGGCACCAATGGGTCG
GAAATCAGTGATAGCGATTATCAGGCAGAATACCCCGATAAATAGCATGGCGAGCACCCAGATGACTACCCTCGAGAGTA
CCAGTATGGCGATGACCAGGGCATTGATTAT
ATGACCCACTTCAACAAGGGCCCTTCCTACGGGCTCTCCGCCGAAGTTAAAGAACAAAATCGCATCCAAGTATGACCAGC
AGGCCGAGGAAGATCTGCCTAACTGGATAGAAGAGGTGACAGGCCTAGGCATTGGCACCAACTACCAGCTGGGGCTGAAG
GACGGCATCATCCTGTGCGAACTCATAAACAAGCTACAGTCAGGCTCTGTGAAAAAAGTCAATGAATCCTCACTAAATTG
GCCGCAGTTGGAGAATATCGGCAACTTCATTAAAGCTATCCAGGCTTACGGTATGAAGCCCCATGATATATTTGAAGCAA
ACGAACTCTTTGAGAATGGCAACATGACCCAGGTTCAGACGACACTGGTGGCTCTAGCAGGTCTGGTGAAAACAAAAGGA
TTCCATACAACCATTGACACTGGTGTTAAGTATGCAGAAAAACAAACAAGACGTTTTGATGAAGGCAAATTAAAGGCTGG
CCAGAGTGTAATTGGTTTACAGATGGGTACCAACAAATGTGCCAGCCAAGCGGGCACGACAGCCTATGGGACTCGGAGGC
ATCTTTATGATCCCAAGATGCAGACGGACAAACCCTTTGACCAGACCACGATTAGCCTGCAGATGGGCACCAACAAAGGA
GCCAGCCAGGCTGGGATGTTAACACCAGGTACCAGAAGAGACATCTATGACCAGAAGTTGACATTACAGCCAGTGGACAA
CTCGACCATTTCTCTACAGATGGGCACCAACAAAACTGCTTCCCAGAAAGGAATGAGCGTGTATTGGCTTGGGCGGCAAG
TGTATGACCCCAAGTACTGTGCCGCACCCACAGAACCTGTCATTCACAACGGAAGCCAGGGCACGGGCACCAATGGGTCG
GAAATCAGTGATAGCGATTATCAGGCAGAATACCCCGATAAATAGCATGGCGAGCACCCAGATGACTACCCTCGAGAGTA
CCAGTATGGCGATGACCAGGGCATTGATTAT
ORF - retro_mmus_1052 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 95.77 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MTHFNKGPSYGLSAEVK-NKIASKYDQQAEEDLRNWIEEVTGLGIGTNFQLGLKDGIILCELINKLQPGS |
| MTHFNKGPSYGLSAEVK.NKIASKYDQQAEEDL.NWIEEVTGLGIGTN.QLGLKDGIILCELINKLQ.GS | |
| Retrocopy | MTHFNKGPSYGLSAEVK>NKIASKYDQQAEEDLPNWIEEVTGLGIGTNYQLGLKDGIILCELINKLQSGS |
| Parental | VKKVNESSLNWPQLENIGNFIKAIQAYGMKPHDIFEANDLFENGNMTQVQTTLVALAGLAKTKGFHTTID |
| VKKVNESSLNWPQLENIGNFIKAIQAYGMKPHDIFEAN.LFENGNMTQVQTTLVALAGL.KTKGFHTTID | |
| Retrocopy | VKKVNESSLNWPQLENIGNFIKAIQAYGMKPHDIFEANELFENGNMTQVQTTLVALAGLVKTKGFHTTID |
| Parental | IGVKYAEKQTRRFDEGKLKAGQSVIGLQMGTNKCASQAGMTAYGTRRHLYDPKMQTDKPFDQTTISLQMG |
| .GVKYAEKQTRRFDEGKLKAGQSVIGLQMGTNKCASQAG.TAYGTRRHLYDPKMQTDKPFDQTTISLQMG | |
| Retrocopy | TGVKYAEKQTRRFDEGKLKAGQSVIGLQMGTNKCASQAGTTAYGTRRHLYDPKMQTDKPFDQTTISLQMG |
| Parental | TNKGASQAGMLAPGTRRDIYDQKLTLQPVDNSTISLQMGTNKVASQKGMSVYGLGRQVYDPKYCAAPTEP |
| TNKGASQAGML.PGTRRDIYDQKLTLQPVDNSTISLQMGTNK.ASQKGMSVY.LGRQVYDPKYCAAPTEP | |
| Retrocopy | TNKGASQAGMLTPGTRRDIYDQKLTLQPVDNSTISLQMGTNKTASQKGMSVYWLGRQVYDPKYCAAPTEP |
| Parental | VIHNGSQGTGTNGSEISDSDYQAEYPDEYHGEYPDDYPREYQYGDDQGIDY |
| VIHNGSQGTGTNGSEISDSDYQAEYPD..HGE.PDDYPREYQYGDDQGIDY | |
| Retrocopy | VIHNGSQGTGTNGSEISDSDYQAEYPDK*HGEHPDDYPREYQYGDDQGIDY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 46 .98 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 53 .36 RPM |
| SRP007412_heart | 0 .03 RPM | 34 .71 RPM |
| SRP007412_kidney | 0 .15 RPM | 122 .12 RPM |
| SRP007412_liver | 0 .08 RPM | 111 .83 RPM |
| SRP007412_testis | 0 .00 RPM | 5 .58 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_1052 was not detected
No EST(s) were mapped for retro_mmus_1052 retrocopy.
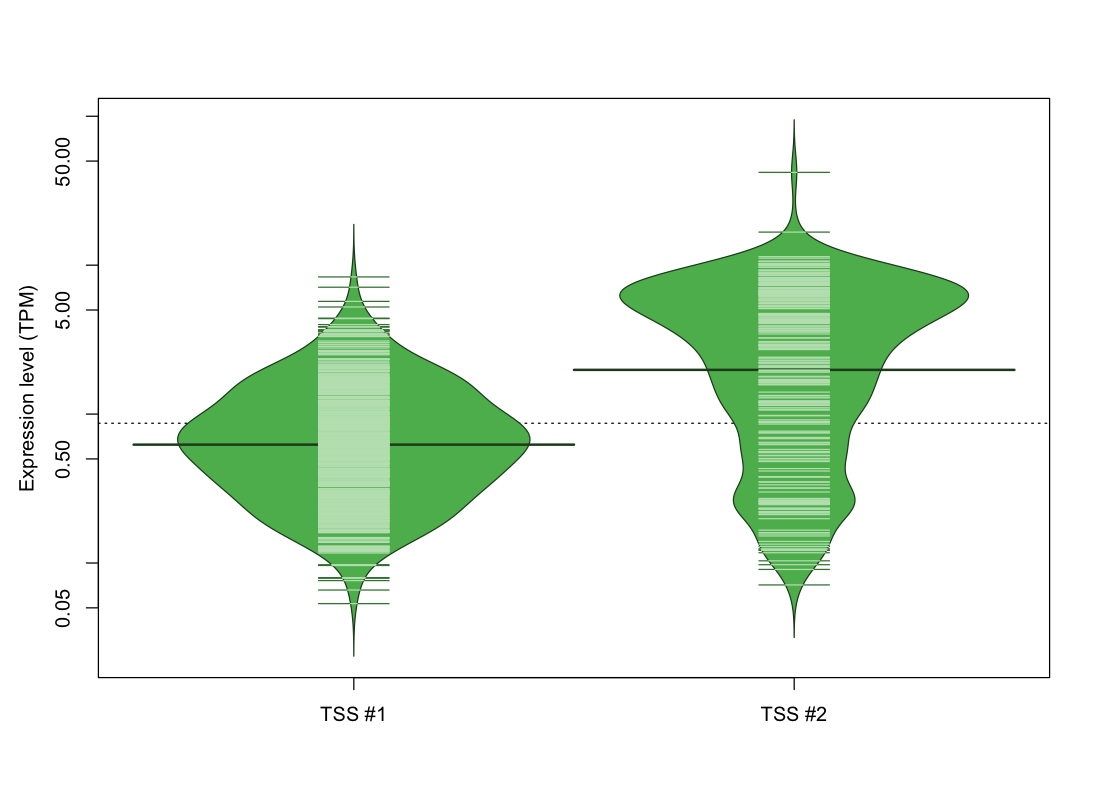
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_31958 | 479 libraries | 419 libraries | 170 libraries | 4 libraries | 0 libraries |
| TSS #2 | TSS_31959 | 834 libraries | 69 libraries | 89 libraries | 70 libraries | 10 libraries |
The graphical summary, for retro_mmus_1052 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1052 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1052 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 13 parental genes, and 19 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000016707 | 1 retrocopy | |
| Bos taurus | ENSBTAG00000020345 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000018128 | 3 retrocopies | |
| Echinops telfairi | ENSETEG00000010095 | 1 retrocopy | |
| Homo sapiens | ENSG00000117519 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000013870 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000002921 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000001690 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000004326 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000004665 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000053931 | 2 retrocopies |
retro_mmus_1052 , retro_mmus_2544,
|
| Sarcophilus harrisii | ENSSHAG00000002670 | 1 retrocopy | |
| Vicugna pacos | ENSVPAG00000004889 | 2 retrocopies |