RetrogeneDB ID: | retro_mmus_132 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 16:36041512..36042499(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000034379 | |
| Aliases: | Wdr5b, 2310009C03Rik, AI606931 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Wdr5 | ||
| Ensembl ID: | ENSMUSG00000026917 | ||
| Aliases: | Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3 | ||
| Description: | WD repeat domain 5 [Source:MGI Symbol;Acc:MGI:2155884] |

Retrocopy-Parental alignment summary:
>retro_mmus_132
ATGGCGACCGAGCATCTGCCAGCAGAGAGAGCACAGTCACTTCTCTCTGCCCCCCGGCGCGAGGAAGAGCCGCAGAAACC
CAACTATGCTCTCAGACTTACTCTTGCGGGGCACTCGGCGGCCATATCATCAGTTAAATTTAGTCCTAATGGCGAATGGC
TAGCGAGTTCTGCAGCTGATGCACTAATTATAATTTGGGGAGCATACGATGGGAATTGTAAGAAAACACTCTATGGTCAC
AGTCTGGAAATATCAGATGTTGCCTGGTCTTCAGATTCTAGTCGTCTAGTTTCAGCCTCAGATGATAAGACTCTAAAGGT
ATGGGATATGAGATCTGGAAAATGTCTGAAAACACTGAAGGGGCACAGCGATTTCGTCTTCTGTTGTGACTTCAACCCAC
CATCCAACCTCATCGTTTCGGGCTCTTTTGATGAGAGTGTGAAAATATGGGAAGTAAAAACAGGAAAATGCCTCAAGACT
TTGTCTGCACATTCGGACCCCATTTCTGCCGTAAATTTTAATTGTAATGGGTCCTTGATAGTGTCCGGGAGCTATGATGG
GCTTTGTAGAATTTGGGATGCTGCGTCAGGCCAGTGTTTAAGGACACTTGCTGATGAAGGCAACCCTCCAGTGTCTTTTG
TAAAATTTTCTCCAAATGGTAAATACATTCTCACTGCAACTTTGGACAATACTCTTAAACTGTGGGATTACAGCAGGGGC
AGATGCCTGAAGACATACACTGGTCATAAGAATGAGAAATACTGCCTATTTGCCAGTTTCTCGGTTACTGGTAGAAAGTG
GGTTGTGTCTGGTTCAGAGGACAACATGGTTTACATCTGGAATCTTCAGACTAAAGAGATTGTACAGAGGTTGCAAGGTC
ACACAGATGTGGTGATCTCAGCAGCCTGTCACCCTACAAAAAACATCATCGCATCAGCAGCACTAGAAAATGACAAGACA
ATTAAAGTGTGGTCGAGTGACTGCTGA
ATGGCGACCGAGCATCTGCCAGCAGAGAGAGCACAGTCACTTCTCTCTGCCCCCCGGCGCGAGGAAGAGCCGCAGAAACC
CAACTATGCTCTCAGACTTACTCTTGCGGGGCACTCGGCGGCCATATCATCAGTTAAATTTAGTCCTAATGGCGAATGGC
TAGCGAGTTCTGCAGCTGATGCACTAATTATAATTTGGGGAGCATACGATGGGAATTGTAAGAAAACACTCTATGGTCAC
AGTCTGGAAATATCAGATGTTGCCTGGTCTTCAGATTCTAGTCGTCTAGTTTCAGCCTCAGATGATAAGACTCTAAAGGT
ATGGGATATGAGATCTGGAAAATGTCTGAAAACACTGAAGGGGCACAGCGATTTCGTCTTCTGTTGTGACTTCAACCCAC
CATCCAACCTCATCGTTTCGGGCTCTTTTGATGAGAGTGTGAAAATATGGGAAGTAAAAACAGGAAAATGCCTCAAGACT
TTGTCTGCACATTCGGACCCCATTTCTGCCGTAAATTTTAATTGTAATGGGTCCTTGATAGTGTCCGGGAGCTATGATGG
GCTTTGTAGAATTTGGGATGCTGCGTCAGGCCAGTGTTTAAGGACACTTGCTGATGAAGGCAACCCTCCAGTGTCTTTTG
TAAAATTTTCTCCAAATGGTAAATACATTCTCACTGCAACTTTGGACAATACTCTTAAACTGTGGGATTACAGCAGGGGC
AGATGCCTGAAGACATACACTGGTCATAAGAATGAGAAATACTGCCTATTTGCCAGTTTCTCGGTTACTGGTAGAAAGTG
GGTTGTGTCTGGTTCAGAGGACAACATGGTTTACATCTGGAATCTTCAGACTAAAGAGATTGTACAGAGGTTGCAAGGTC
ACACAGATGTGGTGATCTCAGCAGCCTGTCACCCTACAAAAAACATCATCGCATCAGCAGCACTAGAAAATGACAAGACA
ATTAAAGTGTGGTCGAGTGACTGCTGA
ORF - retro_mmus_132 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 81.79 % |
| Parental protein coverage: | 93.71 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | SATQSKPTPVKPNYALKFTLAGHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS |
| SA......P.KPNYAL..TLAGH..A.SSVKFSPNGEWLASS.AD.LI.IWGAYDG...KT..GH.L.IS | |
| Retrocopy | SAPRREEEPQKPNYALRLTLAGHSAAISSVKFSPNGEWLASSAADALIIIWGAYDGNCKKTLYGHSLEIS |
| Parental | DVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG |
| DVAWSSDS..LVSASDDKTLK.WD..SGKCLKTLKGHS..VFCC.FNP.SNLIVSGSFDESV.IW.VKTG | |
| Retrocopy | DVAWSSDSSRLVSASDDKTLKVWDMRSGKCLKTLKGHSDFVFCCDFNPPSNLIVSGSFDESVKIWEVKTG |
| Parental | KCLKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILA |
| KCLKTL.AHSDP.SAV.FN..GSLIVS.SYDGLCRIWD.ASGQCL.TL.D..NPPVSFVKFSPNGKYIL. | |
| Retrocopy | KCLKTLSAHSDPISAVNFNCNGSLIVSGSYDGLCRIWDAASGQCLRTLADEGNPPVSFVKFSPNGKYILT |
| Parental | ATLDNTLKLWDYSKGKCLKTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQKLQGHT |
| ATLDNTLKLWDYS.G.CLKTYTGHKNEKYC.FA.FSVTG.KW.VSGSEDN.VYIWNLQTKEIVQ.LQGHT | |
| Retrocopy | ATLDNTLKLWDYSRGRCLKTYTGHKNEKYCLFASFSVTGRKWVVSGSEDNMVYIWNLQTKEIVQRLQGHT |
| Parental | DVVISTACHPTENIIASAALENDKTIKLWKSDC |
| DVVIS.ACHPT.NIIASAALENDKTIK.W.SDC | |
| Retrocopy | DVVISAACHPTKNIIASAALENDKTIKVWSSDC |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 2 .03 RPM | 27 .15 RPM |
| SRP007412_cerebellum | 2 .26 RPM | 32 .61 RPM |
| SRP007412_heart | 1 .62 RPM | 29 .76 RPM |
| SRP007412_kidney | 1 .73 RPM | 41 .69 RPM |
| SRP007412_liver | 0 .71 RPM | 32 .91 RPM |
| SRP007412_testis | 0 .87 RPM | 44 .23 RPM |
RNA Polymerase II actvity may be related with retro_mmus_132 in 4 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 16:36040809..36041673 |
| ENCFF001YIJ | POLR2A | 16:36040263..36042051 |
| ENCFF001YJZ | POLR2A | 16:36040275..36045345 |
| ENCFF001YKA | POLR2A | 16:36039216..36045422 |
1 EST(s) were mapped to retro_mmus_132 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA510402 | 36041631 | 36041878 | 99.6 | 246 | 1 | 245 |
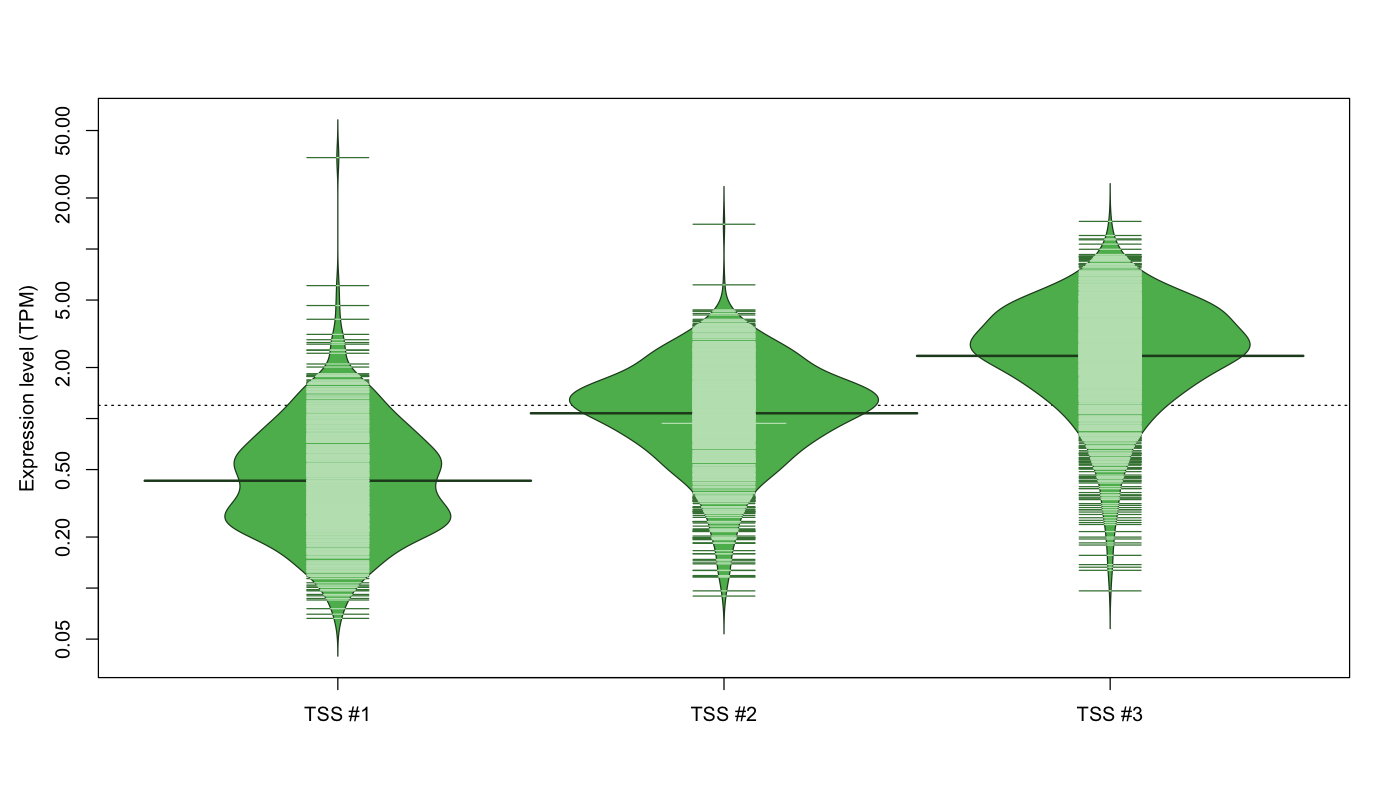
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_46955 | 519 libraries | 477 libraries | 74 libraries | 1 library | 1 library |
| TSS #2 | TSS_46956 | 222 libraries | 336 libraries | 512 libraries | 1 library | 1 library |
| TSS #3 | TSS_46957 | 93 libraries | 119 libraries | 729 libraries | 126 libraries | 5 libraries |
The graphical summary, for retro_mmus_132 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_132 was not experimentally validated.