RetrogeneDB ID: | retro_mmus_1489 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 16:13779819..13780163(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tceb2 | ||
| Ensembl ID: | ENSMUSG00000055839 | ||
| Aliases: | Tceb2, 0610040H15Rik | ||
| Description: | transcription elongation factor B (SIII), polypeptide 2 [Source:MGI Symbol;Acc:MGI:1914923] |

Retrocopy-Parental alignment summary:
>retro_mmus_1489
ATGGCTGTGTTTCTCATGACCCCACACAAGAAGACCACCATCTTCACTGAGGCCAAGGAGTCAAGCTTGGTGTTGGCACT
CAAGCGCATCATTGAGGGCATCATGAAGAGGCCTCCTGAGGAGAGCTGCTGTTCAAGGACAACCAGCTCCTTGATGAGAA
AACTCTGGGGGAATGTGGCTTCACTAGTCAGGTAGCAAAGCCACAGGCCCCAGCCACAGTGGGCCTGGCCTTCAGAGCAG
AAGATGCCTTTGAAGGCCTGCCTATCTACCCCAATTCTTGTCCTCCAGAACTTCCAGGTGTGATGAAGGATTCTGGAAAT
AGTGCTAAGGAACAAGTTGTACAG
ATGGCTGTGTTTCTCATGACCCCACACAAGAAGACCACCATCTTCACTGAGGCCAAGGAGTCAAGCTTGGTGTTGGCACT
CAAGCGCATCATTGAGGGCATCATGAAGAGGCCTCCTGAGGAGAGCTGCTGTTCAAGGACAACCAGCTCCTTGATGAGAA
AACTCTGGGGGAATGTGGCTTCACTAGTCAGGTAGCAAAGCCACAGGCCCCAGCCACAGTGGGCCTGGCCTTCAGAGCAG
AAGATGCCTTTGAAGGCCTGCCTATCTACCCCAATTCTTGTCCTCCAGAACTTCCAGGTGTGATGAAGGATTCTGGAAAT
AGTGCTAAGGAACAAGTTGTACAG
ORF - retro_mmus_1489 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 72.27 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MDVFLMIRRHKTTIFTDAKESSTVFELKRIVEGILKRPPEE-QRLYKDDQLLDDGKTLGECGFTSQTARP |
| M.VFLM....KTTIFT.AKESS.V..LKRI.EGI.KRPPEE...L.KD.QLLD..KTLGECGFTSQ.A.P | |
| Retrocopy | MAVFLMTPHKKTTIFTEAKESSLVLALKRIIEGIMKRPPEE<ELLFKDNQLLDE-KTLGECGFTSQVAKP |
| Parental | QAPATVGLAFRADDTFEALRIEPFSSPPELPDVMKPQDSGGSANEQAVQ |
| QAPATVGLAFRA.D.FE.L.I.P.S.PPELP.VMK..DSG.SA.EQ.VQ | |
| Retrocopy | QAPATVGLAFRAEDAFEGLPIYPNSCPPELPGVMK--DSGNSAKEQVVQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .19 RPM | 34 .18 RPM |
| SRP007412_cerebellum | 0 .04 RPM | 20 .36 RPM |
| SRP007412_heart | 0 .12 RPM | 27 .80 RPM |
| SRP007412_kidney | 0 .19 RPM | 19 .50 RPM |
| SRP007412_liver | 0 .05 RPM | 14 .88 RPM |
| SRP007412_testis | 0 .14 RPM | 71 .72 RPM |
RNA Polymerase II actvity may be related with retro_mmus_1489 in 3 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 16:13780449..13781011 |
| ENCFF001YIJ | POLR2A | 16:13779945..13781469 |
| ENCFF001YKA | POLR2A | 16:13779151..13782127 |
No EST(s) were mapped for retro_mmus_1489 retrocopy.
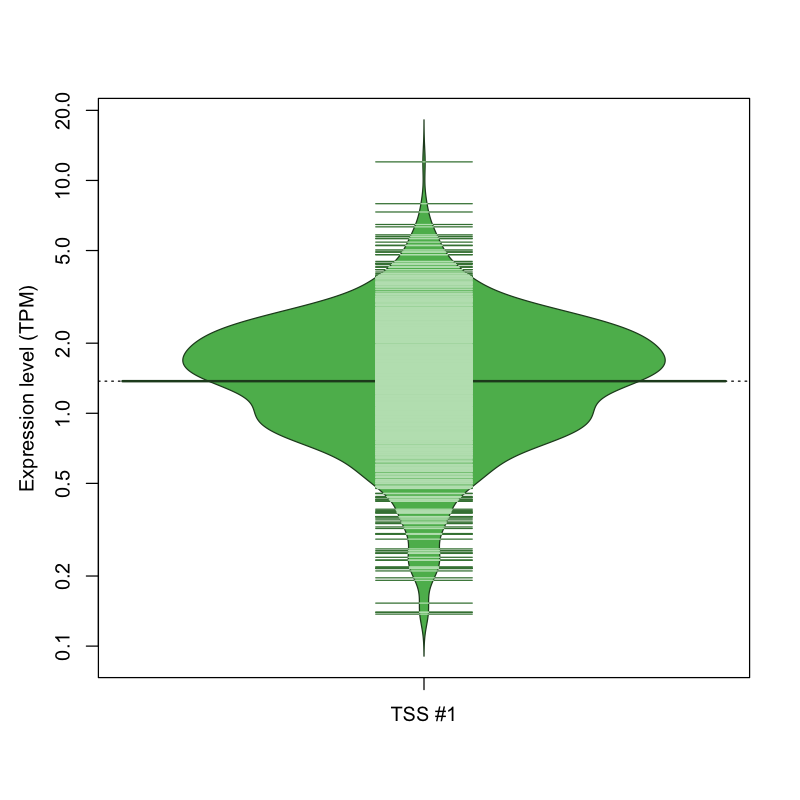
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_44943 | 156 libraries | 265 libraries | 638 libraries | 12 libraries | 1 library |
The graphical summary, for retro_mmus_1489 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1489 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1489 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 15 parental genes, and 44 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000019543 | 3 retrocopies | |
| Callithrix jacchus | ENSCJAG00000015347 | 10 retrocopies | |
| Cavia porcellus | ENSCPOG00000021024 | 1 retrocopy | |
| Felis catus | ENSFCAG00000004711 | 2 retrocopies | |
| Homo sapiens | ENSG00000103363 | 2 retrocopies | |
| Gorilla gorilla | ENSGGOG00000015306 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000016536 | 4 retrocopies | |
| Mustela putorius furo | ENSMPUG00000015265 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000055839 | 7 retrocopies |
retro_mmus_1212, retro_mmus_1489 , retro_mmus_1664, retro_mmus_2930, retro_mmus_511, retro_mmus_747, retro_mmus_99,
|
| Nomascus leucogenys | ENSNLEG00000009471 | 2 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000010116 | 2 retrocopies | |
| Pongo abelii | ENSPPYG00000007013 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000007663 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000004814 | 6 retrocopies | |
| Sus scrofa | ENSSSCG00000008061 | 1 retrocopy |