RetrogeneDB ID: | retro_mmus_80 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 8:92901484..92902228(+) | ||
| Located in intron of: | ENSMUSG00000033192 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000078144 | |
| Aliases: | Capns2, 2310005G05Rik, 30K-2 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Capns1 | ||
| Ensembl ID: | ENSMUSG00000001794 | ||
| Aliases: | Capns1, Capa-4, Capa4, Capn4, D7Ertd146e | ||
| Description: | calpain, small subunit 1 [Source:MGI Symbol;Acc:MGI:88266] |

Retrocopy-Parental alignment summary:
>retro_mmus_80
ATGTTTCTCGCAAAGGCGATCCTGGAAGGTGCAGATCGGGGGCTTGGAGGGGCTCTTGGAGGCCTTCTTGGAGGAGGTGG
TCAGGCAAGAGCAGGCGGGGGCAACATCGGGGGAATCCTTGGAGGAATTGTCAATTTTATCAGCGAGGCTGCCGCAGCTC
AGTACACCCCAGAGCCGCCTCCCCAACAGCAGCATTTTACCGTCGTGGAGGCCTCAGAAAGTGAGGAAGTCAGACGTTTT
CGGCAGCAATTTACACAGCTGGCTGGACCCGACATGGAGGTGGGTGCTACTGACCTGATGAATATTCTCAACAAAGTCCT
TTCTAAGCATAAAGAGCTGAAGACAGAGGGCTTCAGCCTTGATACCTGTAGGAGCATTGTGTCTGTCATGGACAGTGACA
CCACAGGGAAACTGGGATTTGAGGAATTTAAGTACCTGTGGAACAACATCAAGAAATGGCAGTGTGTTTTCAAGCAGTAC
GACAGTGACCATTCTGGCTCCCTCGGAAGCTCCCAGCTGCACGGGGCCATGCAGGCAGCGGGCTTCCAGCTCAATGAGCA
ACTTTACCTAATGATTGTCCGTCGGTATGCTGACGAAGATGGCGGGATGGATTTTAACAACTTTATCAGCTGCCTGGTTC
GCCTGGATGCTATGTTTCGAGCTTTCAAGGCTCTGGACCGAGATAGGGATGGCCTGATTCAGGTGTCCATCCGAGAATGG
CTGCAGCTGACCATGTATTCCTGA
ATGTTTCTCGCAAAGGCGATCCTGGAAGGTGCAGATCGGGGGCTTGGAGGGGCTCTTGGAGGCCTTCTTGGAGGAGGTGG
TCAGGCAAGAGCAGGCGGGGGCAACATCGGGGGAATCCTTGGAGGAATTGTCAATTTTATCAGCGAGGCTGCCGCAGCTC
AGTACACCCCAGAGCCGCCTCCCCAACAGCAGCATTTTACCGTCGTGGAGGCCTCAGAAAGTGAGGAAGTCAGACGTTTT
CGGCAGCAATTTACACAGCTGGCTGGACCCGACATGGAGGTGGGTGCTACTGACCTGATGAATATTCTCAACAAAGTCCT
TTCTAAGCATAAAGAGCTGAAGACAGAGGGCTTCAGCCTTGATACCTGTAGGAGCATTGTGTCTGTCATGGACAGTGACA
CCACAGGGAAACTGGGATTTGAGGAATTTAAGTACCTGTGGAACAACATCAAGAAATGGCAGTGTGTTTTCAAGCAGTAC
GACAGTGACCATTCTGGCTCCCTCGGAAGCTCCCAGCTGCACGGGGCCATGCAGGCAGCGGGCTTCCAGCTCAATGAGCA
ACTTTACCTAATGATTGTCCGTCGGTATGCTGACGAAGATGGCGGGATGGATTTTAACAACTTTATCAGCTGCCTGGTTC
GCCTGGATGCTATGTTTCGAGCTTTCAAGGCTCTGGACCGAGATAGGGATGGCCTGATTCAGGTGTCCATCCGAGAATGG
CTGCAGCTGACCATGTATTCCTGA
ORF - retro_mmus_80 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 67.77 % |
| Parental protein coverage: | 88.81 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | LISGAAGGGGGGGGGMGLGGGG---GGGGTAMRILGGVISAISEAAA-QYNPEPPPPRSHYSNIEANESE |
| ...GA..G.GG..GG..LGGGG....GGG....ILGG....ISEAAA.QY.PEPPP...H....EA.ESE | |
| Retrocopy | ILEGADRGLGGALGGL-LGGGGQARAGGGNIGGILGGIVNFISEAAAAQYTPEPPPQQQHFTVVEASESE |
| Parental | EVRQFRKLFVQLAGDDMEVSATELMNILNKVVTRHPDLKTDGFGIDTCRSMVAVMDSDTTGKLGFEEFKY |
| EVR.FR..F.QLAG.DMEV.AT.LMNILNKV...H..LKT.GF..DTCRS.V.VMDSDTTGKLGFEEFKY | |
| Retrocopy | EVRRFRQQFTQLAGPDMEVGATDLMNILNKVLSKHKELKTEGFSLDTCRSIVSVMDSDTTGKLGFEEFKY |
| Parental | LWNNIKKWQAIYKRFDTDRSGTIGSHELPGAFEAAGFHLNEHLYSMIIRRYADESGNMDFDNFISCLVRL |
| LWNNIKKWQ...K..D.D.SG..GS..L.GA..AAGF.LNE.LY.MI.RRYADE.G.MDF.NFISCLVRL | |
| Retrocopy | LWNNIKKWQCVFKQYDSDHSGSLGSSQLHGAMQAAGFQLNEQLYLMIVRRYADEDGGMDFNNFISCLVRL |
| Parental | DAMFRAFKSLDKNGTGQIQVNIQEWLQLTMYS |
| DAMFRAFK.LD....G.IQV.I.EWLQLTMYS | |
| Retrocopy | DAMFRAFKALDRDRDGLIQVSIREWLQLTMYS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .40 RPM | 111 .60 RPM |
| SRP007412_cerebellum | 0 .04 RPM | 161 .31 RPM |
| SRP007412_heart | 0 .00 RPM | 265 .70 RPM |
| SRP007412_kidney | 0 .06 RPM | 196 .98 RPM |
| SRP007412_liver | 0 .03 RPM | 77 .75 RPM |
| SRP007412_testis | 0 .91 RPM | 188 .17 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_80 was not detected
1 EST(s) were mapped to retro_mmus_80 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| EH109772 | 92901645 | 92901921 | 100 | 276 | 0 | 276 |
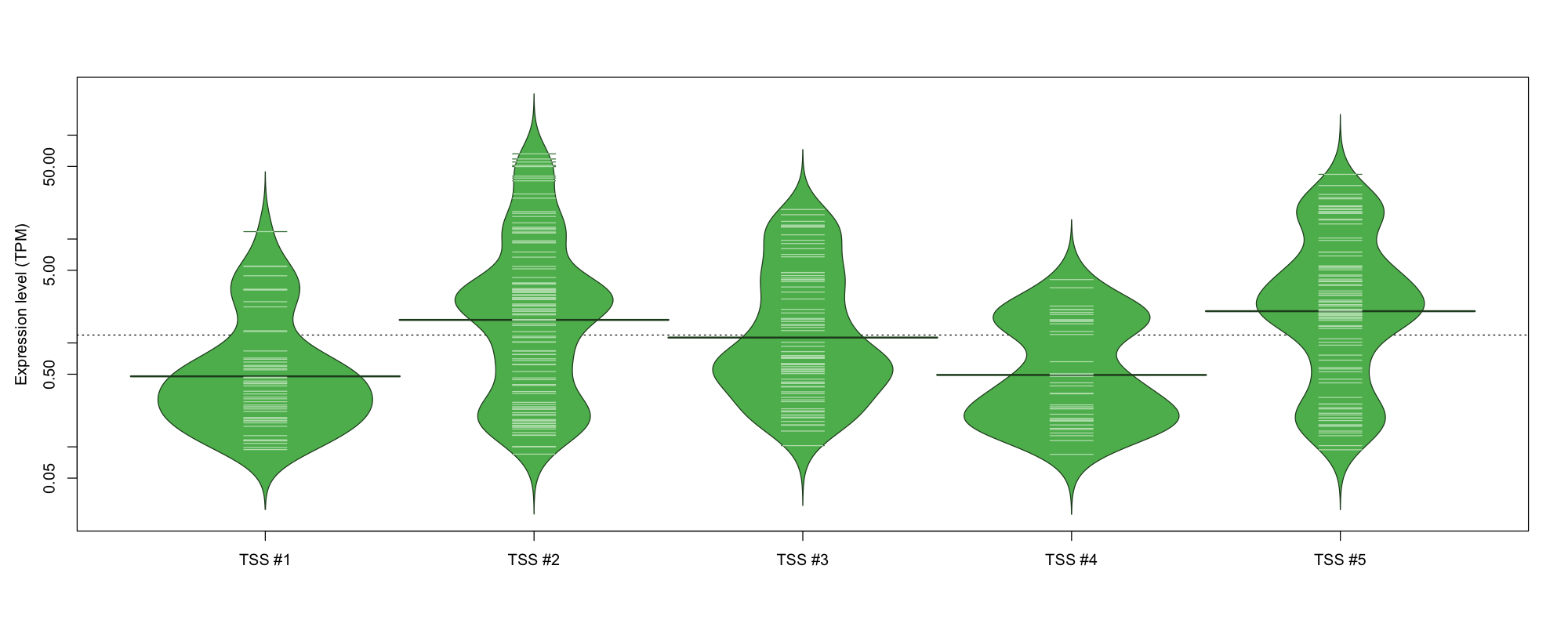
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_145333 | 1022 libraries | 39 libraries | 8 libraries | 2 libraries | 1 library |
| TSS #2 | TSS_145334 | 959 libraries | 43 libraries | 43 libraries | 7 libraries | 20 libraries |
| TSS #3 | TSS_145335 | 996 libraries | 43 libraries | 20 libraries | 5 libraries | 8 libraries |
| TSS #4 | TSS_145336 | 1033 libraries | 25 libraries | 14 libraries | 0 libraries | 0 libraries |
| TSS #5 | TSS_145337 | 984 libraries | 26 libraries | 37 libraries | 8 libraries | 17 libraries |
The graphical summary, for retro_mmus_80 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_80 was not experimentally validated.