RetrogeneDB ID: | retro_mmus_134 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:50599219..50600410(+) | ||
| Located in intron of: | ENSMUSG00000055409 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000084234 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Idh3g | ||
| Ensembl ID: | ENSMUSG00000002010 | ||
| Aliases: | None | ||
| Description: | isocitrate dehydrogenase 3 (NAD+), gamma [Source:MGI Symbol;Acc:MGI:1099463] |

Retrocopy-Parental alignment summary:
>retro_mmus_134
ATGTTGGCAGTAACCAGCTGTTCTATGAAAACAGTTCTCCAATATGCTGTGTTTCTTGGCCATTCTAGGGAGGTAGTGTG
TGAGCTTGTGACCTCCTTCAGAAGTTTCTGCTCACATTGCGCAGTGCCTCCTTCACCAAAGTATGGAGGAAGACACACGG
TGGCTATGATTCCTGGAGATGGCATTGGGCCTGAGCTTATGGTTCATGTCAAGAAAATATTCAGATCAAATTGTGTGCCA
GTGGACTTTGAAGAGGTGTGGGTCACCTCGACATCTAATGAAGAGGAGATCAACAATGCTCTCATGGCCATCCGTCGGAA
CCGTGTCGCTCTGAAGGGCAATATTGCAACCAACCACAATCTGCCTGCCCGTTACAAGTCTCACAACACCAAGTTTCGCA
CCATCCTTGACCTCTATGCCAGTGTCGTCCATTTCAAGACTTTTCCTGGTGTTATGACCCGACACAAAGACATAGACATC
TTGGTTGTTCGGGAGAATACAGAGGGGGAGTACACTAACCTAGAGCATGAGAGCGTGAAAGGCGTGGTAGAAAGCCTTAA
GATTGTGACAAAGACCAAGTCTGTGCGCATTGCTGATTATGCCTTCAAGTTGGCCCAAAAGATGGGGCGGAAAAAGGTGA
CAGTTGTTCACAAAGCCAACATCATGAAACTGGGAGATGGGCTCTTCCTTCAGTGCTGTAAGGATGTGGCAGCTCACTAT
CCTCAGATCACCTTAGAGAGTATGATTATAGACAACACCACAATGCAGTTGGTATCGAAGCCCCAACAGTTTGATGTCAT
GGTGATGCCCAATCTTTATGGCAACATTATCAACAGTATCTGCACAGGACTGGTTGGAGGGTCAGGAATTGTCCCTGGAG
CCAATTATGGTGATTCATATGCAATATTTGAGATGGGTTCAAAGGAAATAGGTAAAGATTTGGCTCACAGAAATATTGCC
AATCCTGTCGCCATGCTGCTGACCAGCTGTATCATGCTGGACTACCTTGATCTCCAACCCTATGCCACTCACATCCGCTC
TGCAGTCATGGCGTCCTTACAAAACAAAGCTGTCTGCACTCCAGACATTGGAGGTCAAGGGAACACTGCAAGCACTGTAG
AGTATATCTTACACCACATGAAAGAACAAACTAGTGGTTGTCATCCAAATTTCTTCCTGCAGTTCACTTGA
ORF - retro_mmus_134 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 69.83 % |
| Parental protein coverage: | 91.09 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | EVLAAHVAPRRSISSQQTIPPSAKYGGRHTVTMIPGDGIGPELMLHVKSVFRHACVPVDFEEVHVSSNAD |
| EV....V...RS..S....PPS.KYGGRHTV.MIPGDGIGPELM.HVK..FR..CVPVDFEEV.V.S... | |
| Retrocopy | EVVCELVTSFRSFCSHCAVPPSPKYGGRHTVAMIPGDGIGPELMVHVKKIFRSNCVPVDFEEVWVTSTSN |
| Parental | EEDIRNAIMAIRRNRVALKGNIETNHNLPPSHKSRNNILRTSLDLYANVIHCKSLPGVVTRHKDIDILIV |
| EE.I.NA.MAIRRNRVALKGNI.TNHNLP...KS.N...RT.LDLYA.V.H.K..PGV.TRHKDIDIL.V | |
| Retrocopy | EEEINNALMAIRRNRVALKGNIATNHNLPARYKSHNTKFRTILDLYASVVHFKTFPGVMTRHKDIDILVV |
| Parental | RENTEGEYSSLEHESVAGVVESLKIITKAKSLRIAEYAFKLAQESGRKKVTAVHKANIMKLGDGLFLQCC |
| RENTEGEY..LEHESV.GVVESLKI.TK.KS.RIA.YAFKLAQ..GRKKVT.VHKANIMKLGDGLFLQCC | |
| Retrocopy | RENTEGEYTNLEHESVKGVVESLKIVTKTKSVRIADYAFKLAQKMGRKKVTVVHKANIMKLGDGLFLQCC |
| Parental | REVAAHYPQITFDSMIVDNTTMQLVSRPQQFDVMVMPNLYGNIVNNVCAGLVGGPGLVAGANYGHVYAVF |
| ..VAAHYPQIT..SMI.DNTTMQLVS.PQQFDVMVMPNLYGNI.N..C.GLVGG.G.V.GANYG..YA.F | |
| Retrocopy | KDVAAHYPQITLESMIIDNTTMQLVSKPQQFDVMVMPNLYGNIINSICTGLVGGSGIVPGANYGDSYAIF |
| Parental | ETATRNTGKSIANKNIANPTATLLASCMMLDHLKLHSYATSIRKAVLASMDNENMHTPDIGGQGTTSQAI |
| E......GK..A..NIANP.A.LL.SC.MLD.L.L..YAT.IR.AV.AS..N....TPDIGGQG.T.... | |
| Retrocopy | EMGSKEIGKDLAHRNIANPVAMLLTSCIMLDYLDLQPYATHIRSAVMASLQNKAVCTPDIGGQGNTASTV |
| Parental | QDIIRHIR |
| ..I..H.. | |
| Retrocopy | EYILHHMK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .21 RPM | 124 .10 RPM |
| SRP007412_cerebellum | 0 .04 RPM | 216 .80 RPM |
| SRP007412_heart | 0 .00 RPM | 490 .61 RPM |
| SRP007412_kidney | 0 .00 RPM | 361 .80 RPM |
| SRP007412_liver | 0 .00 RPM | 65 .08 RPM |
| SRP007412_testis | 37 .92 RPM | 16 .42 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_134 was not detected
1 EST(s) were mapped to retro_mmus_134 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| BB616954 | 50599189 | 50599841 | 99 | 644 | 7 | 636 |
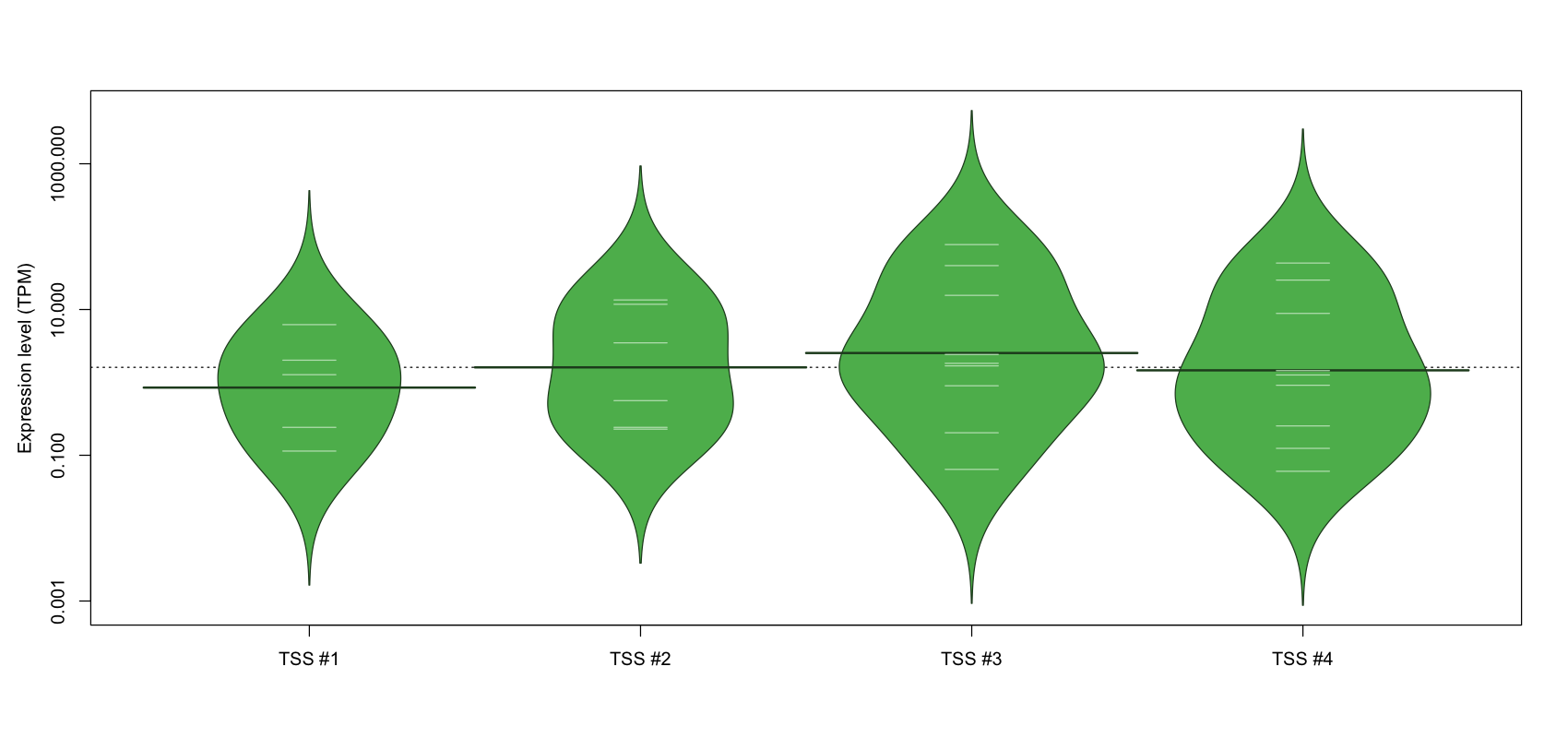
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_136205 | 1067 libraries | 2 libraries | 2 libraries | 1 library | 0 libraries |
| TSS #2 | TSS_136206 | 1066 libraries | 3 libraries | 1 library | 0 libraries | 2 libraries |
| TSS #3 | TSS_136207 | 1063 libraries | 3 libraries | 3 libraries | 0 libraries | 3 libraries |
| TSS #4 | TSS_136208 | 1063 libraries | 4 libraries | 2 libraries | 1 library | 2 libraries |
The graphical summary, for retro_mmus_134 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_134 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_134 has 7 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 18 parental genes, and 18 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000018466 | 1 retrocopy | |
| Bos taurus | ENSBTAG00000001059 | 1 retrocopy | |
| Canis familiaris | ENSCAFG00000019286 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000011185 | 1 retrocopy | |
| Equus caballus | ENSECAG00000018509 | 1 retrocopy | |
| Homo sapiens | ENSG00000067829 | 1 retrocopy | |
| Latimeria chalumnae | ENSLACG00000007365 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000006904 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000013934 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000004457 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000008781 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000002010 | 1 retrocopy |
retro_mmus_134 ,
|
| Nomascus leucogenys | ENSNLEG00000013540 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000006735 | 1 retrocopy | |
| Pteropus vampyrus | ENSPVAG00000013163 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000037284 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000012783 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000011728 | 1 retrocopy |