RetrogeneDB ID: | retro_hsap_3005 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 4:165109535..165111485(+) | ||
| Located in intron of: | ENSG00000145416 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000250746 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | HSPA5 | ||
| Ensembl ID: | ENSG00000044574 | ||
| Aliases: | HSPA5, BIP, GRP78, HEL-S-89n, MIF2 | ||
| Description: | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) [Source:HGNC Symbol;Acc:5238] |

Retrocopy-Parental alignment summary:
>retro_hsap_3005
ATGAAGCTCTCCCTGGTGGCGGCGATGCTGCTGCTACTCGGTGAGGCGTGGTTCGAGGACAAGGAGGAGGACGTGGGCAC
GGTGGTCTGCGTCGACCTGGGGACCACCTACTGCTGCGTCTGTGTGTTCAAGACCGGCTGCGTGGAGATCACCGCCAACA
ATCAGAGCAAACGCATCACGCGGTCCTATGTGCCCTTCACTCTTGAAGGGGAATATCTGATCGGCGATGCCGCCACGAAC
CAGCTCACCTCCAATCCCAAGAACACGGTCTTTGACGCCAAGTGGCTCATCGGCCGCACGTGGAACGACCTGTCTATGCA
GCAGGACATCAAGTTCTTGCGGTTCAAGGTAGTTGAAAAGAAAACTAAACCATACGTTCAAGTTGATATTGGAGGTGGGC
AAACAAAGACATTTGCTCCTGAAGAAATTTCTGCCATGGTTCTCACTAAAATGCAATAAACCGCTGAGGCTTATTTGAGA
AAGAAGTTTACCCATGCAGTTTTTACCTTACCAGCCTATTTCAATGATGCCCAATGCGAAGCAACCAAAGATATTGGAAC
TATTGCTGAGCTAAATGTTATGAGGATCATTAACGAGCCTATGGCAGCTGCTATTGGTTATGGCCTGGATAAGAGGGAGG
GGGAGAAGACATCCTGTTGTTCCACCTGGGTGGCAGAACCTTCAATGCGTCTCCTCTCACCATTGACAATGGTGTCTTCG
GAGTCGTGGCCACTAATGGAGATACTCATCTGCGTGCAGAAGACTATGACTAGCGTGTCATGGAACATTTCATCAAGTTG
TACAAAAAGAAGACTGACAAAGATGTCAGGAAAGACGATAGGGCTGTGCAGAAACTCCAGCTAGAGGTAGAAAAGGCCAG
ATGGGCCCTGTCTCCTCAACATCAAGCAAGAATTGAAATTGAGTCCTTCTATGAAGGAGAAGACTTTTCTGAGACCCTGA
CTCAGGCCAAATTTTAAGAGCTAAACATGGATCTGTTCTGGTCTACTATGAGGCCTGTCCAGAAAGTGTTGGAAAATTCT
GATTTGAAGAATTCTGATATTGATGATGTTGTTCTTGGTGGCTCTACTCGAATTCCAAAGATTCAGCAACTGGTTAAAAA
GTCCTTCAATGGCAAGGAGTCCTCCCATGGCATAAACCCAGATGAAGCTGTAGTGCATGGTGCTGCTGTCCAGGCTGGAG
TGCTCTCTGGTGATCAAGATACAGGTGACCTGGTAGTGCTTGATGTATGTCCCCTTACATTTGGTATTGAAACTGTGGGA
GGTGTCATGATCAAACTGATTCCAAGGAACACAGTGGTACCTACCAAGAAGTCCATATCTTTTCTATAGCTTCTGATAAT
CAACCAACTGTCATAATCAAGGTCTATGAGGATGAATGACCCCTGACAAAAGACATTCATCTTCTGGGTACATTTGATCT
GACTGGAATTCCTCCTGCTCCTCGTGGGGTCCCACAGATTGAAGTCACCTTTGAGATAGATGTGAATGGTATTCTTCGAG
TGACAGCTGAAGACAAGGGTACAGGGAACAAAAATAAGATCACAATTACCAATGACCAGAATCACCTGACACCTGAAGAA
ATCAAAAGGATGGTTAATGATGCTGAGAAGTTTGCTGAGGAAGACACAAAGCTCAAGGAGTGCATTGATACTACAAATGA
GCTGGAAAGCTATGGCTATTCTCTAAAGAATCAGATTGGAGATAAAGAAAAGCTGGGAGGTAAACTTTCCTCTGAAGATA
AGGAGACCATGGAAAAAGCTGTAGAAGAAAAGATTGAATGGCTGGAAAGCCACCAAGCTGCTGACATTGAAGACTTCGAA
GCTAACAAGAAGGAACTGGAAGAAATTGTTCAGCCACTTATCAGCAAACTCTATGGAAGTGCAAGCCCTCTCCCAGCCAC
GAAGAGGATACGGCAGAAAAAGATGAGTTG
ATGAAGCTCTCCCTGGTGGCGGCGATGCTGCTGCTACTCGGTGAGGCGTGGTTCGAGGACAAGGAGGAGGACGTGGGCAC
GGTGGTCTGCGTCGACCTGGGGACCACCTACTGCTGCGTCTGTGTGTTCAAGACCGGCTGCGTGGAGATCACCGCCAACA
ATCAGAGCAAACGCATCACGCGGTCCTATGTGCCCTTCACTCTTGAAGGGGAATATCTGATCGGCGATGCCGCCACGAAC
CAGCTCACCTCCAATCCCAAGAACACGGTCTTTGACGCCAAGTGGCTCATCGGCCGCACGTGGAACGACCTGTCTATGCA
GCAGGACATCAAGTTCTTGCGGTTCAAGGTAGTTGAAAAGAAAACTAAACCATACGTTCAAGTTGATATTGGAGGTGGGC
AAACAAAGACATTTGCTCCTGAAGAAATTTCTGCCATGGTTCTCACTAAAATGCAATAAACCGCTGAGGCTTATTTGAGA
AAGAAGTTTACCCATGCAGTTTTTACCTTACCAGCCTATTTCAATGATGCCCAATGCGAAGCAACCAAAGATATTGGAAC
TATTGCTGAGCTAAATGTTATGAGGATCATTAACGAGCCTATGGCAGCTGCTATTGGTTATGGCCTGGATAAGAGGGAGG
GGGAGAAGACATCCTGTTGTTCCACCTGGGTGGCAGAACCTTCAATGCGTCTCCTCTCACCATTGACAATGGTGTCTTCG
GAGTCGTGGCCACTAATGGAGATACTCATCTGCGTGCAGAAGACTATGACTAGCGTGTCATGGAACATTTCATCAAGTTG
TACAAAAAGAAGACTGACAAAGATGTCAGGAAAGACGATAGGGCTGTGCAGAAACTCCAGCTAGAGGTAGAAAAGGCCAG
ATGGGCCCTGTCTCCTCAACATCAAGCAAGAATTGAAATTGAGTCCTTCTATGAAGGAGAAGACTTTTCTGAGACCCTGA
CTCAGGCCAAATTTTAAGAGCTAAACATGGATCTGTTCTGGTCTACTATGAGGCCTGTCCAGAAAGTGTTGGAAAATTCT
GATTTGAAGAATTCTGATATTGATGATGTTGTTCTTGGTGGCTCTACTCGAATTCCAAAGATTCAGCAACTGGTTAAAAA
GTCCTTCAATGGCAAGGAGTCCTCCCATGGCATAAACCCAGATGAAGCTGTAGTGCATGGTGCTGCTGTCCAGGCTGGAG
TGCTCTCTGGTGATCAAGATACAGGTGACCTGGTAGTGCTTGATGTATGTCCCCTTACATTTGGTATTGAAACTGTGGGA
GGTGTCATGATCAAACTGATTCCAAGGAACACAGTGGTACCTACCAAGAAGTCCATATCTTTTCTATAGCTTCTGATAAT
CAACCAACTGTCATAATCAAGGTCTATGAGGATGAATGACCCCTGACAAAAGACATTCATCTTCTGGGTACATTTGATCT
GACTGGAATTCCTCCTGCTCCTCGTGGGGTCCCACAGATTGAAGTCACCTTTGAGATAGATGTGAATGGTATTCTTCGAG
TGACAGCTGAAGACAAGGGTACAGGGAACAAAAATAAGATCACAATTACCAATGACCAGAATCACCTGACACCTGAAGAA
ATCAAAAGGATGGTTAATGATGCTGAGAAGTTTGCTGAGGAAGACACAAAGCTCAAGGAGTGCATTGATACTACAAATGA
GCTGGAAAGCTATGGCTATTCTCTAAAGAATCAGATTGGAGATAAAGAAAAGCTGGGAGGTAAACTTTCCTCTGAAGATA
AGGAGACCATGGAAAAAGCTGTAGAAGAAAAGATTGAATGGCTGGAAAGCCACCAAGCTGCTGACATTGAAGACTTCGAA
GCTAACAAGAAGGAACTGGAAGAAATTGTTCAGCCACTTATCAGCAAACTCTATGGAAGTGCAAGCCCTCTCCCAGCCAC
GAAGAGGATACGGCAGAAAAAGATGAGTTG
ORF - retro_hsap_3005 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 84.78 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 4 |
| Number of frameshifts detected | 3 |
Retrocopy - Parental Gene Alignment:
| Parental | MKLSLVAAMLLLLSAARAEEEDKKEDVGTVVGIDLGTTYSCVGVFKNGRVEIIANDQGNRITPSYVAFTP |
| MKLSLVAAMLLLL..A..E..DK.EDVGTVV..DLGTTY.CV.VFK.G.VEI.AN.Q..RIT.SYV.FT. | |
| Retrocopy | MKLSLVAAMLLLLGEAWFE--DKEEDVGTVVCVDLGTTYCCVCVFKTGCVEITANNQSKRITRSYVPFTL |
| Parental | EGERLIGDAAKNQLTSNPENTVFDAKRLIGRTWNDPSVQQDIKFLPFKVVEKKTKPYIQVDIGGGQTKTF |
| EGE.LIGDAA.NQLTSNP.NTVFDAK.LIGRTWND.S.QQDIKFL.FKVVEKKTKPY.QVDIGGGQTKTF | |
| Retrocopy | EGEYLIGDAATNQLTSNPKNTVFDAKWLIGRTWNDLSMQQDIKFLRFKVVEKKTKPYVQVDIGGGQTKTF |
| Parental | APEEISAMVLTKMKETAEAYLGKKVTHAVVTVPAYFNDAQRQATKDAGTIAGLNVMRIINEPTAAAIAYG |
| APEEISAMVLTKM..TAEAYL.KK.THAV.T.PAYFNDAQ..ATKD.GTIA.LNVMRIINEP.AAAI.YG | |
| Retrocopy | APEEISAMVLTKMQ*TAEAYLRKKFTHAVFTLPAYFNDAQCEATKDIGTIAELNVMRIINEPMAAAIGYG |
| Parental | LDKREGEK-NILVFDLGGGTFDVSLLTIDNGVFEVVATNGDTHLGGEDFDQRVMEHFIKLYKKKTGKDVR |
| LDKREGEK..IL.F.LGG.TF..S.LTIDNGVF.VVATNGDTHL..ED.D.RVMEHFIKLYKKKT.KDVR | |
| Retrocopy | LDKREGEK<DILLFHLGGRTFNASPLTIDNGVFGVVATNGDTHLRAEDYD*RVMEHFIKLYKKKTDKDVR |
| Parental | KDNRAVQKLRREVEKAKRALSSQHQARIEIESFYEGEDFSETLTRAKFEELNMDLFRSTMKPVQKVLEDS |
| KD.RAVQKL..EVEKA..ALS.QHQARIEIESFYEGEDFSETLT.AKF.ELNMDLF.STM.PVQKVLE.S | |
| Retrocopy | KDDRAVQKLQLEVEKARWALSPQHQARIEIESFYEGEDFSETLTQAKF*ELNMDLFWSTMRPVQKVLENS |
| Parental | DLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDEAVAYGAAVQAGVLSGDQDTGDLVLLDV |
| DLK.SDID..VL.GGSTRIPKIQQLVK..FNGKE.S.GINPDEAV..GAAVQAGVLSGDQDTGDLV.LDV | |
| Retrocopy | DLKNSDIDDVVL-GGSTRIPKIQQLVKKSFNGKESSHGINPDEAVVHGAAVQAGVLSGDQDTGDLVVLDV |
| Parental | CPLTLGIETVGGVMTKLIPRNTVVPTKKS-QIFSTASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIP |
| CPLT.GIETVGGVM.KLIPRNTVVPTKKS..IFS.ASDNQPTV.IKVYE.E.PLTKD.HLLGTFDLTGIP | |
| Retrocopy | CPLTFGIETVGGVMIKLIPRNTVVPTKKS<HIFSIASDNQPTVIIKVYEDE*PLTKDIHLLGTFDLTGIP |
| Parental | PAPRGVPQIEVTFEIDVNGILRVTAEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKER |
| PAPRGVPQIEVTFEIDVNGILRVTAEDKGTGNKNKITITNDQN.LTPEEI.RMVNDAEKFAEED.KLKE. | |
| Retrocopy | PAPRGVPQIEVTFEIDVNGILRVTAEDKGTGNKNKITITNDQNHLTPEEIKRMVNDAEKFAEEDTKLKEC |
| Parental | IDTRNELESYAYSLKNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQ |
| IDT.NELESY.YSLKNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQ.ADIEDF.A.KKELEEIVQ | |
| Retrocopy | IDTTNELESYGYSLKNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQAADIEDFEANKKELEEIVQ |
| Parental | PIISKLYGSAG-PPPTGEEDTAEKDEL |
| P.ISKLYGSA..P.P..EEDTAEKDEL | |
| Retrocopy | PLISKLYGSAS<PSPSHEEDTAEKDEL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 285 .92 RPM |
| bodymap2_adrenal | 0 .00 RPM | 487 .99 RPM |
| bodymap2_brain | 0 .05 RPM | 172 .10 RPM |
| bodymap2_breast | 0 .00 RPM | 293 .97 RPM |
| bodymap2_colon | 0 .02 RPM | 1067 .92 RPM |
| bodymap2_heart | 0 .02 RPM | 237 .28 RPM |
| bodymap2_kidney | 0 .00 RPM | 1434 .23 RPM |
| bodymap2_liver | 0 .00 RPM | 950 .08 RPM |
| bodymap2_lung | 0 .02 RPM | 1331 .80 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 940 .07 RPM |
| bodymap2_ovary | 0 .06 RPM | 594 .23 RPM |
| bodymap2_prostate | 0 .00 RPM | 352 .91 RPM |
| bodymap2_skeletal_muscle | 0 .04 RPM | 161 .45 RPM |
| bodymap2_testis | 0 .00 RPM | 677 .67 RPM |
| bodymap2_thyroid | 0 .00 RPM | 308 .63 RPM |
| bodymap2_white_blood_cells | 0 .02 RPM | 364 .89 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_3005 was not detected
No EST(s) were mapped for retro_hsap_3005 retrocopy.
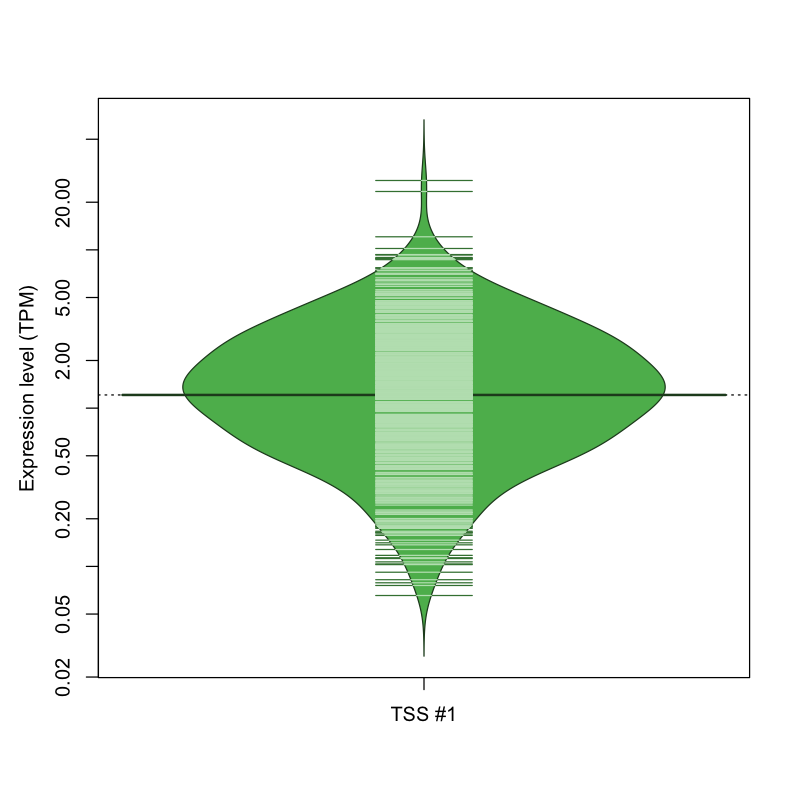
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_141425 | 1190 libraries | 253 libraries | 347 libraries | 35 libraries | 4 libraries |
The graphical summary, for retro_hsap_3005 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_3005 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_3005 has 4 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_2033 |
| Gorilla gorilla | retro_ggor_2082 |
| Pongo abelii | retro_pabe_2491 |
| Callithrix jacchus | retro_cjac_2221 |
Parental genes homology:
Parental genes homology involve 13 parental genes, and 23 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000006880 | 1 retrocopy | |
| Homo sapiens | ENSG00000044574 | 1 retrocopy |
retro_hsap_3005 ,
|
| Homo sapiens | ENSG00000109971 | 8 retrocopies | |
| Homo sapiens | ENSG00000113013 | 1 retrocopy | |
| Gallus gallus | ENSGALG00000001000 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000026879 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000016702 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000026864 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000017348 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000006786 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000019609 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000000563 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000011243 | 4 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .03 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .04 RPM |
| CEU_NA12400 | 0 .04 RPM |
| CEU_NA12751 | 0 .05 RPM |
| CEU_NA12760 | 0 .04 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .03 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .03 RPM |
| FIN_HG00277 | 0 .04 RPM |
| FIN_HG00315 | 0 .11 RPM |
| FIN_HG00321 | 0 .03 RPM |
| FIN_HG00328 | 0 .02 RPM |
| FIN_HG00338 | 0 .06 RPM |
| FIN_HG00349 | 0 .14 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .10 RPM |
| FIN_HG00378 | 0 .04 RPM |
| GBR_HG00099 | 0 .03 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .02 RPM |
| GBR_HG00131 | 0 .03 RPM |
| GBR_HG00133 | 0 .02 RPM |
| GBR_HG00134 | 0 .02 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .03 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .03 RPM |
| TSI_NA20513 | 0 .02 RPM |
| TSI_NA20518 | 0 .08 RPM |
| TSI_NA20532 | 0 .07 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .03 RPM |
| TSI_NA20765 | 0 .08 RPM |
| TSI_NA20771 | 0 .03 RPM |
| TSI_NA20786 | 0 .03 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .03 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .02 RPM |
| YRI_NA19093 | 0 .03 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .05 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .02 RPM |