RetrogeneDB ID: | retro_mmus_1826 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 19:37784265..37785584(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Psmc1 | ||
| Ensembl ID: | ENSMUSG00000021178 | ||
| Aliases: | Psmc1, AI325227, P26s4, S4 | ||
| Description: | protease (prosome, macropain) 26S subunit, ATPase 1 [Source:MGI Symbol;Acc:MGI:106054] |

Retrocopy-Parental alignment summary:
>retro_mmus_1826
ATGGGTCAAAGTCAGAGTGGTGGCCATGGTCCTGGGGGTGGCAAGAAGGATGACAAGGACAAGAAAAAGGAATATGAACC
TCCTGTCCCAACTAGAGTGGGGAAAAAGAAGAAGAAAACGAAGGGACCAGCTGCTGCCAGCAAACTTCCACTGGTAACAA
CTCACACCCAGTGCCACCTGAAATTACTAAAGCTAGAGAGAATAAAAGACTATCTTCTCATGGAGGAGGAATTCATTAGA
AATCAGGAACAGATGAAACCATTAGAAGAAAAGCAAGAAGAGGAAAGATCAAAAATGGATGATCCCAGGGGGACCCCCAT
GTCTGTAGGAACCTTGGAAGAGATCATTGATGATAATCATGCCATTGTGTCCACATCTGTGGGCTCAGAACACTACAGTC
AGCATCCTGCCATTTGTAGACAAGGATCTGCTGGAACCAGGCTGTTCAGTCCTGCTCAACCACAAGGTGCATGCTGTGAT
AGGGGTGCTAATGGATGACACGGATCCCCTGATCACGGTGATGAAGGTGGAAAAGGACCCCCAGGAAACCTATGCAGATA
TTGGGGGACTGGACAACCAGATCCAAGAAATTAAGGAGTCTGTGGAGCTTGCTCTTACCCATCCTGGCTATTACGAAGAG
ATGGGGATAAAGCCCCCTAAGGGGGTCATTCTCTACGGTCCACCAGGAACAGGTAAAGCTCTATTGGCCAAAGCAGTAGC
GAACCAGACTTCAGCCATTCTCTTGCGAGTGGTTGGCTCAGAGCTTATCCAGAAGTATGTAGGTGACGCGGCCCAAACTT
GTCCGGGAGCTCTTCCGGGTTGCTGAGGAGCACACACCGTCCATCGTGTTCATTGATGAGATCGACACCATTGGGACCAA
AAGATATGATTCAAACTCTGGAGGTGAGAGAGAAATTCGACGAACAATGTTGGAACTGTTGAACCAGTTGGATGGATTTG
ATTCAAGGGGAGATGTAAAAGTTATCACGGCCACAAACCAAATAGAAACTTTGGATCCAGCACTTATCAGGCCAGGCCCC
ATTGACAGAAAGATCGAGTTCCCCCTGCCTGATGAGACCAAGAAGCATATCTTCCAGATTCACACCAGCAGGATGACACT
GGCTGATGATGTAACCTTGGATGACTTGATCATGGCAAAGGATGACCTCTCTGGGGCCGACATCAAGGCAATCTGTACAG
AAGCTGGCTTGATGGCCTTACGGGGACGCAGAATGAAAGTAACAAATGAAGACTACAAAAAATCTAAAGAGAATGTTCTT
TATAAAAAACAAGAAGGCACCCCCAAGGGATTCGGTCTC
ATGGGTCAAAGTCAGAGTGGTGGCCATGGTCCTGGGGGTGGCAAGAAGGATGACAAGGACAAGAAAAAGGAATATGAACC
TCCTGTCCCAACTAGAGTGGGGAAAAAGAAGAAGAAAACGAAGGGACCAGCTGCTGCCAGCAAACTTCCACTGGTAACAA
CTCACACCCAGTGCCACCTGAAATTACTAAAGCTAGAGAGAATAAAAGACTATCTTCTCATGGAGGAGGAATTCATTAGA
AATCAGGAACAGATGAAACCATTAGAAGAAAAGCAAGAAGAGGAAAGATCAAAAATGGATGATCCCAGGGGGACCCCCAT
GTCTGTAGGAACCTTGGAAGAGATCATTGATGATAATCATGCCATTGTGTCCACATCTGTGGGCTCAGAACACTACAGTC
AGCATCCTGCCATTTGTAGACAAGGATCTGCTGGAACCAGGCTGTTCAGTCCTGCTCAACCACAAGGTGCATGCTGTGAT
AGGGGTGCTAATGGATGACACGGATCCCCTGATCACGGTGATGAAGGTGGAAAAGGACCCCCAGGAAACCTATGCAGATA
TTGGGGGACTGGACAACCAGATCCAAGAAATTAAGGAGTCTGTGGAGCTTGCTCTTACCCATCCTGGCTATTACGAAGAG
ATGGGGATAAAGCCCCCTAAGGGGGTCATTCTCTACGGTCCACCAGGAACAGGTAAAGCTCTATTGGCCAAAGCAGTAGC
GAACCAGACTTCAGCCATTCTCTTGCGAGTGGTTGGCTCAGAGCTTATCCAGAAGTATGTAGGTGACGCGGCCCAAACTT
GTCCGGGAGCTCTTCCGGGTTGCTGAGGAGCACACACCGTCCATCGTGTTCATTGATGAGATCGACACCATTGGGACCAA
AAGATATGATTCAAACTCTGGAGGTGAGAGAGAAATTCGACGAACAATGTTGGAACTGTTGAACCAGTTGGATGGATTTG
ATTCAAGGGGAGATGTAAAAGTTATCACGGCCACAAACCAAATAGAAACTTTGGATCCAGCACTTATCAGGCCAGGCCCC
ATTGACAGAAAGATCGAGTTCCCCCTGCCTGATGAGACCAAGAAGCATATCTTCCAGATTCACACCAGCAGGATGACACT
GGCTGATGATGTAACCTTGGATGACTTGATCATGGCAAAGGATGACCTCTCTGGGGCCGACATCAAGGCAATCTGTACAG
AAGCTGGCTTGATGGCCTTACGGGGACGCAGAATGAAAGTAACAAATGAAGACTACAAAAAATCTAAAGAGAATGTTCTT
TATAAAAAACAAGAAGGCACCCCCAAGGGATTCGGTCTC
ORF - retro_mmus_1826 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 92.99 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MGQSQSGGHGPGGGKKDDKDKKKKYEPPVPTRVGKKKKKTKGPDAASKLPLVTPHTQCRLKLLKLERIKD |
| MGQSQSGGHGPGGGKKDDKDKKK.YEPPVPTRVGKKKKKTKGP.AASKLPLVT.HTQC.LKLLKLERIKD | |
| Retrocopy | MGQSQSGGHGPGGGKKDDKDKKKEYEPPVPTRVGKKKKKTKGPAAASKLPLVTTHTQCHLKLLKLERIKD |
| Parental | YLLMEEEFIRNQEQMKPLEEKQEEERSKVDDLRGTPMSVGTLEEIIDDNHAIVSTSVGSEHY-VSILSFV |
| YLLMEEEFIRNQEQMKPLEEKQEEERSK.DD.RGTPMSVGTLEEIIDDNHAIVSTSVGSEHY.VSIL.FV | |
| Retrocopy | YLLMEEEFIRNQEQMKPLEEKQEEERSKMDDPRGTPMSVGTLEEIIDDNHAIVSTSVGSEHY>VSILPFV |
| Parental | DKDLLEPGCSVLLNHKVHAVIGVLMDDTDPLVTVMKVEKAPQETYADIGGLDNQIQEIKESVELPLTHPE |
| DKDLLEPGCSVLLNHKVHAVIGVLMDDTDPL.TVMKVEK.PQETYADIGGLDNQIQEIKESVEL.LTHP. | |
| Retrocopy | DKDLLEPGCSVLLNHKVHAVIGVLMDDTDPLITVMKVEKDPQETYADIGGLDNQIQEIKESVELALTHPG |
| Parental | YYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELIQKYLGDG-PKLVRELFRVAEEHA |
| YYEEMGIKPPKGVILYGPPGTGK.LLAKAVANQTSA..LRVVGSELIQKY.GD..PKLVRELFRVAEEH. | |
| Retrocopy | YYEEMGIKPPKGVILYGPPGTGKALLAKAVANQTSAILLRVVGSELIQKYVGDA>PKLVRELFRVAEEHT |
| Parental | PSIVFIDEIDAIGTKRYDSNSGGEREIQRTMLELLNQLDGFDSRGDVKVIMATNRIETLDPALIRPGRID |
| PSIVFIDEID.IGTKRYDSNSGGEREI.RTMLELLNQLDGFDSRGDVKVI.ATN.IETLDPALIRPG.ID | |
| Retrocopy | PSIVFIDEIDTIGTKRYDSNSGGEREIRRTMLELLNQLDGFDSRGDVKVITATNQIETLDPALIRPGPID |
| Parental | RKIEFPLPDEKTKKRIFQIHTSRMTLADDVTLDDLIMAKDDLSGADIKAICTEAGLMALRERRMKVTNED |
| RKIEFPLPDE.TKK.IFQIHTSRMTLADDVTLDDLIMAKDDLSGADIKAICTEAGLMALR.RRMKVTNED | |
| Retrocopy | RKIEFPLPDE-TKKHIFQIHTSRMTLADDVTLDDLIMAKDDLSGADIKAICTEAGLMALRGRRMKVTNED |
| Parental | FKKSKENVLYKKQEGTPEGLYL |
| .KKSKENVLYKKQEGTP.G..L | |
| Retrocopy | YKKSKENVLYKKQEGTPKGFGL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 96 .00 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 109 .38 RPM |
| SRP007412_heart | 0 .09 RPM | 87 .29 RPM |
| SRP007412_kidney | 0 .08 RPM | 101 .66 RPM |
| SRP007412_liver | 0 .00 RPM | 96 .81 RPM |
| SRP007412_testis | 0 .09 RPM | 60 .28 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_1826 was not detected
No EST(s) were mapped for retro_mmus_1826 retrocopy.
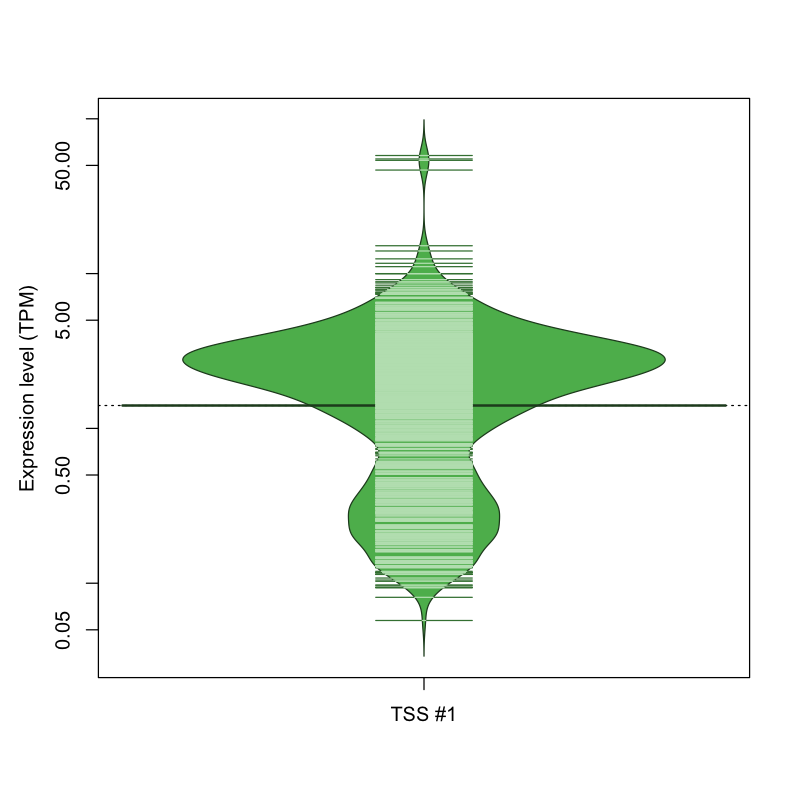
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_64288 | 319 libraries | 234 libraries | 455 libraries | 55 libraries | 9 libraries |
The graphical summary, for retro_mmus_1826 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1826 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1826 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 17 parental genes, and 48 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000001839 | 4 retrocopies | |
| Canis familiaris | ENSCAFG00000017510 | 2 retrocopies | |
| Choloepus hoffmanni | ENSCHOG00000013595 | 1 retrocopy | |
| Dasypus novemcinctus | ENSDNOG00000006458 | 1 retrocopy | |
| Equus caballus | ENSECAG00000003601 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000002661 | 1 retrocopy | |
| Felis catus | ENSFCAG00000011310 | 8 retrocopies | |
| Macropus eugenii | ENSMEUG00000001526 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000007471 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000022990 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000008150 | 4 retrocopies | |
| Mus musculus | ENSMUSG00000021178 | 3 retrocopies |
retro_mmus_1517, retro_mmus_1826 , retro_mmus_812,
|
| Oryctolagus cuniculus | ENSOCUG00000006231 | 4 retrocopies | |
| Otolemur garnettii | ENSOGAG00000032343 | 2 retrocopies | |
| Pongo abelii | ENSPPYG00000025948 | 9 retrocopies | |
| Pteropus vampyrus | ENSPVAG00000010020 | 3 retrocopies | |
| Tursiops truncatus | ENSTTRG00000013582 | 1 retrocopy |