RetrogeneDB ID: | retro_mmus_3308 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 9:119162624..119163405(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ywhaq | ||
| Ensembl ID: | ENSMUSG00000076432 | ||
| Aliases: | Ywhaq, 2700028P07Rik, AA409740, AU021156, R74690 | ||
| Description: | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide [Source:MGI Symbol;Acc:MGI:891963] |

Retrocopy-Parental alignment summary:
>retro_mmus_3308
ACTCGCGTCGCGGCCGCGGAGACTAGAAGGAGGACTCCGGATCCGGCTCGGCGCTTGCCCTCGCTCGCCATGGAGACCGA
GCTGATCTAGAAGGCCAAGCTGGCCGACATGGCCACCTGCATGAAAGCTGTGACAGAGCAGGGCGCCGAGCTGTCCAACG
AGGAGCACAACCTGCTGTCGGTGGCCTACAAAAACGTGGTAGGGGGCCGCAGGTCCGCCTGGAGGGTCATCTCGAGCATT
GAGCAGAAGACCGACACCTCTGACAAGACGTTGCAGCTGATCAAGGACTAATGGGGGAAAGTGGAATCGGAGCTGAGGTC
CATCTGCACCATGGTCCTGGAATTGTTGGATAAGTATTTGATAGCCAATGCAACTAATCCAGAGAGTAAGGTCTTCTATC
TGAACATGAAGGGAGATTATTTCCGGTATCTTGCTGAAGTAGCTTGTGGCGATGATCGAAAACAAACAATAGAAAATTCC
CAAGGAGCCTACCAAGAGGCATTTGATATAAGCAAGAAGGAGATGCAGCCTACGCATCTGATCCGCCTGGGGCTGGCTCT
TAACTTTTCTGTATTTTACTATGAGATCCTTAATAATCCAGAGCTTGCCTGCACACTGGCTAAAACGGCTTTTGATGAGG
CCATCACAGAGCTTGATACACTGAACGAAGACTCCTACAAAGACAGCACCCTCATCATGCAGTTGCTTAGAGACAACTTA
ACATTATGGACATCAGACAGTGCAGGGAGAAGAATGTGATGCAGCAGAGGGGGCCGAAAAC
ACTCGCGTCGCGGCCGCGGAGACTAGAAGGAGGACTCCGGATCCGGCTCGGCGCTTGCCCTCGCTCGCCATGGAGACCGA
GCTGATCTAGAAGGCCAAGCTGGCCGACATGGCCACCTGCATGAAAGCTGTGACAGAGCAGGGCGCCGAGCTGTCCAACG
AGGAGCACAACCTGCTGTCGGTGGCCTACAAAAACGTGGTAGGGGGCCGCAGGTCCGCCTGGAGGGTCATCTCGAGCATT
GAGCAGAAGACCGACACCTCTGACAAGACGTTGCAGCTGATCAAGGACTAATGGGGGAAAGTGGAATCGGAGCTGAGGTC
CATCTGCACCATGGTCCTGGAATTGTTGGATAAGTATTTGATAGCCAATGCAACTAATCCAGAGAGTAAGGTCTTCTATC
TGAACATGAAGGGAGATTATTTCCGGTATCTTGCTGAAGTAGCTTGTGGCGATGATCGAAAACAAACAATAGAAAATTCC
CAAGGAGCCTACCAAGAGGCATTTGATATAAGCAAGAAGGAGATGCAGCCTACGCATCTGATCCGCCTGGGGCTGGCTCT
TAACTTTTCTGTATTTTACTATGAGATCCTTAATAATCCAGAGCTTGCCTGCACACTGGCTAAAACGGCTTTTGATGAGG
CCATCACAGAGCTTGATACACTGAACGAAGACTCCTACAAAGACAGCACCCTCATCATGCAGTTGCTTAGAGACAACTTA
ACATTATGGACATCAGACAGTGCAGGGAGAAGAATGTGATGCAGCAGAGGGGGCCGAAAAC
ORF - retro_mmus_3308 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 85.88 % |
| Parental protein coverage: | 86.14 % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | TRRRTPDPARRSPSLAMEKTELIQKAKLAEQAERYDDMATCMKAVTEQGAELSNEERNLLSVAYKNVVGG |
| TR.......RR.P..A.....L.....L...A....DMATCMKAVTEQGAELSNEE.NLLSVAYKNVVGG | |
| Retrocopy | TRVAAAETRRRTPDPARRLPSLAMETELI*KA-KLADMATCMKAVTEQGAELSNEEHNLLSVAYKNVVGG |
| Parental | RRSAWRVISSIEQKTDTSDKKLQLIKDYREKVESELRSICTTVLELLDKYLIANATNPESKVFYLKMKGD |
| RRSAWRVISSIEQKTDTSDK.LQLIKD...KVESELRSICT.VLELLDKYLIANATNPESKVFYL.MKGD | |
| Retrocopy | RRSAWRVISSIEQKTDTSDKTLQLIKD*WGKVESELRSICTMVLELLDKYLIANATNPESKVFYLNMKGD |
| Parental | YFRYLAEVACGDDRKQTIENSQGAYQEAFDISKKEMQPTHPIRLGLALNFSVFYYEILNNPELACTLAKT |
| YFRYLAEVACGDDRKQTIENSQGAYQEAFDISKKEMQPTH.IRLGLALNFSVFYYEILNNPELACTLAKT | |
| Retrocopy | YFRYLAEVACGDDRKQTIENSQGAYQEAFDISKKEMQPTHLIRLGLALNFSVFYYEILNNPELACTLAKT |
| Parental | AFDEAIAELDTLNEDSYKDSTLIMQLLRDNLTLWTSDSA-GEECDAAEGAEN |
| AFDEAI.ELDTLNEDSYKDSTLIMQLLRDNLTLWTSDSA.GEECDAAEGAEN | |
| Retrocopy | AFDEAITELDTLNEDSYKDSTLIMQLLRDNLTLWTSDSA>GEECDAAEGAEN |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .16 RPM | 71 .61 RPM |
| SRP007412_cerebellum | 0 .17 RPM | 42 .60 RPM |
| SRP007412_heart | 0 .00 RPM | 9 .62 RPM |
| SRP007412_kidney | 0 .04 RPM | 23 .25 RPM |
| SRP007412_liver | 0 .03 RPM | 10 .16 RPM |
| SRP007412_testis | 0 .32 RPM | 33 .98 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_3308 was not detected
No EST(s) were mapped for retro_mmus_3308 retrocopy.
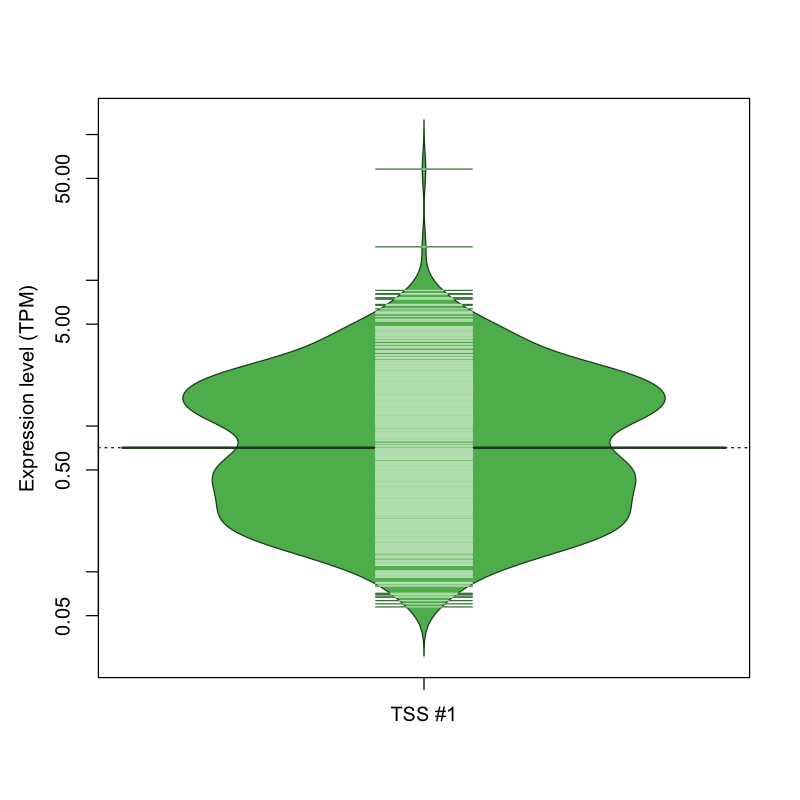
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_147604 | 388 libraries | 391 libraries | 274 libraries | 17 libraries | 2 libraries |
The graphical summary, for retro_mmus_3308 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3308 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3308 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 13 parental genes, and 42 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Equus caballus | ENSECAG00000004925 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000015359 | 4 retrocopies | |
| Felis catus | ENSFCAG00000007854 | 2 retrocopies | |
| Myotis lucifugus | ENSMLUG00000005383 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000014740 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000022285 | 3 retrocopies | |
| Mus musculus | ENSMUSG00000076432 | 10 retrocopies |
retro_mmus_1076, retro_mmus_1378, retro_mmus_1926, retro_mmus_2250, retro_mmus_2299, retro_mmus_2471, retro_mmus_2689, retro_mmus_3250, retro_mmus_3308 , retro_mmus_406,
|
| Nomascus leucogenys | ENSNLEG00000013534 | 3 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000010482 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000000547 | 3 retrocopies | |
| Rattus norvegicus | ENSRNOG00000008104 | 5 retrocopies | |
| Sorex araneus | ENSSARG00000012476 | 4 retrocopies | |
| Tursiops truncatus | ENSTTRG00000000871 | 4 retrocopies |