RetrogeneDB ID: | retro_mmus_338 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:77065966..77067101(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tfdp1 | ||
| Ensembl ID: | ENSMUSG00000038482 | ||
| Aliases: | Tfdp1, Dp1, Drtf1 | ||
| Description: | transcription factor Dp 1 [Source:MGI Symbol;Acc:MGI:101934] |

Retrocopy-Parental alignment summary:
>retro_mmus_338
GTGGCGTCCCTTGTCACCATCCACCGTTCCACAGTGAACACACTTGGAAAGCAGCTTTTGCCAAAAACCTTCGGACAGTC
CAATGTCAATGTCACACAGCAAGTGGTGATTGGCACGCCTCACAGTCCTGCAGCATCCAACACTATTGTGGTAGGGAGCC
CACACTCTCCCAACACTCACTTTGTGTCCCAGAATCAGACGTCTGACTCCTCACCTTGGTCTGCTGGGAGGTGGAACAGG
AAAGGCGAGAAGAATAGCAAGGGCCTGTGGCATTTCTCCATGAAGGTGTGTGAGAAGGTGCAGAGGAAAGGAACTACCTC
CTACAGTGAGGTGGCTGACGAGCTGGTGGCAGAGTTCAGTACTGCCGACAACCACATCCTACCAAACCAATCAGCTTATG
ACCAGAAGAACATCTGGAGGAGTGTCTATGATGCCTTAAATGTGCTAATAACCATGAACATCATCTTCAAGGAGAAGAAG
GAGAGCAAATGGATCGGCCTGCCCACCAACTCAGCTCAGGAGTGCCAGAACTTAGAGGTAGAGAGGCAGCGGAGGCTGGA
GAGGATCAAACAGAAGCAGTCACAGCTCCAGGAGCTCATCTGCAGCAAATTGCCTTCAAGAACTTGGTGCAGAGAAATCA
CCAAGCTGAGCAGCAGGCCTGCAGGCTGTCTCCTCCCACCTCTGTCATCCACTTGCCCTTCATCATTGTCAACACCAGCA
GGAAGACAGTCATTGACTGCAGCATCTCCAATGACAAATTTGAGTATCTGTTTAACTTTGACAACACGTTTGAAATCCAT
GATGACATTGAGGTGCTCAAGTGCATGGGCACTGCGTGTGGGCTGGAGTGTGGCAACTGCTCTGCTGAAGACCTCAAGGT
GGTGAGAAGTTTGGTACTAAAAGCTCTAGAACCATAAGTGACAGAAATGGCTCAGGGATCCATTGGTGGAGTATTCCTCA
CGACAACAGGTTCTACATCCAATGGCACAAGGCTTTCTGCCAGTGATTTGAGCAATGGTGCAGATGGAATGCTGGCCATA
AGCTCCATGGGTCTCAGTACAGAGGCTCCAGGGTTGAGAGCCCTGTGTCCTACATTGGGGAGGATGATGACAACAATGAC
TATGACTTTAATGAG
GTGGCGTCCCTTGTCACCATCCACCGTTCCACAGTGAACACACTTGGAAAGCAGCTTTTGCCAAAAACCTTCGGACAGTC
CAATGTCAATGTCACACAGCAAGTGGTGATTGGCACGCCTCACAGTCCTGCAGCATCCAACACTATTGTGGTAGGGAGCC
CACACTCTCCCAACACTCACTTTGTGTCCCAGAATCAGACGTCTGACTCCTCACCTTGGTCTGCTGGGAGGTGGAACAGG
AAAGGCGAGAAGAATAGCAAGGGCCTGTGGCATTTCTCCATGAAGGTGTGTGAGAAGGTGCAGAGGAAAGGAACTACCTC
CTACAGTGAGGTGGCTGACGAGCTGGTGGCAGAGTTCAGTACTGCCGACAACCACATCCTACCAAACCAATCAGCTTATG
ACCAGAAGAACATCTGGAGGAGTGTCTATGATGCCTTAAATGTGCTAATAACCATGAACATCATCTTCAAGGAGAAGAAG
GAGAGCAAATGGATCGGCCTGCCCACCAACTCAGCTCAGGAGTGCCAGAACTTAGAGGTAGAGAGGCAGCGGAGGCTGGA
GAGGATCAAACAGAAGCAGTCACAGCTCCAGGAGCTCATCTGCAGCAAATTGCCTTCAAGAACTTGGTGCAGAGAAATCA
CCAAGCTGAGCAGCAGGCCTGCAGGCTGTCTCCTCCCACCTCTGTCATCCACTTGCCCTTCATCATTGTCAACACCAGCA
GGAAGACAGTCATTGACTGCAGCATCTCCAATGACAAATTTGAGTATCTGTTTAACTTTGACAACACGTTTGAAATCCAT
GATGACATTGAGGTGCTCAAGTGCATGGGCACTGCGTGTGGGCTGGAGTGTGGCAACTGCTCTGCTGAAGACCTCAAGGT
GGTGAGAAGTTTGGTACTAAAAGCTCTAGAACCATAAGTGACAGAAATGGCTCAGGGATCCATTGGTGGAGTATTCCTCA
CGACAACAGGTTCTACATCCAATGGCACAAGGCTTTCTGCCAGTGATTTGAGCAATGGTGCAGATGGAATGCTGGCCATA
AGCTCCATGGGTCTCAGTACAGAGGCTCCAGGGTTGAGAGCCCTGTGTCCTACATTGGGGAGGATGATGACAACAATGAC
TATGACTTTAATGAG
ORF - retro_mmus_338 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 88.71 % |
| Parental protein coverage: | 92.2 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | VVSLVAVHPSTVNTLGKQLLPKTFGQSNVNITQQVVIGTPQRPAASNTIVVGSPHTPNTHFVSQNQTSDS |
| V.SLV..H.STVNTLGKQLLPKTFGQSNVN.TQQVVIGTP..PAASNTIVVGSPH.PNTHFVSQNQTSDS | |
| Retrocopy | VASLVTIHRSTVNTLGKQLLPKTFGQSNVNVTQQVVIGTPHSPAASNTIVVGSPHSPNTHFVSQNQTSDS |
| Parental | SPWSAGKRNRKGEKNGKGLRHFSMKVCEKVQRKGTTSYNEVADELVAEFSAADNHILPNESAYDQKNIRR |
| SPWSAG..NRKGEKN.KGL.HFSMKVCEKVQRKGTTSY.EVADELVAEFS.ADNHILPN.SAYDQKNI.R | |
| Retrocopy | SPWSAGRWNRKGEKNSKGLWHFSMKVCEKVQRKGTTSYSEVADELVAEFSTADNHILPNQSAYDQKNIWR |
| Parental | RVYDALNVLMAMNIISKEKKEIKWIGLPTNSAQECQNLEVERQRRLERIKQKQSQLQELI-LQQIAFKNL |
| .VYDALNVL..MNII.KEKKE.KWIGLPTNSAQECQNLEVERQRRLERIKQKQSQLQELI.LQQIAFKNL | |
| Retrocopy | SVYDALNVLITMNIIFKEKKESKWIGLPTNSAQECQNLEVERQRRLERIKQKQSQLQELI<LQQIAFKNL |
| Parental | VQRNRQAEQQARRPPPPNSVIHLPFIIVNTSRKTVIDCSISNDKFEYLFNFDNTFEIHDDIEVLKRMGMA |
| VQRN.QAEQQA.R..PP.SVIHLPFIIVNTSRKTVIDCSISNDKFEYLFNFDNTFEIHDDIEVLK.MG.A | |
| Retrocopy | VQRNHQAEQQACRLSPPTSVIHLPFIIVNTSRKTVIDCSISNDKFEYLFNFDNTFEIHDDIEVLKCMGTA |
| Parental | CGLESGNCSAEDLKVARSLVPKALEPYVTEMAQGSIGGVFVTTTGSTSNGTRLSASDLSNGADGMLATSS |
| CGLE.GNCSAEDLKV.RSLV.KALEP.VTEMAQGSIGGVF.TTTGSTSNGTRLSASDLSNGADGMLA.SS | |
| Retrocopy | CGLECGNCSAEDLKVVRSLVLKALEP*VTEMAQGSIGGVFLTTTGSTSNGTRLSASDLSNGADGMLAISS |
| Parental | -NGSQYSGSRVETPVSYVGEDDDDDD-DFNE |
| ..GSQY.GSRVE.PVSY.GEDDD..D.DFNE | |
| Retrocopy | <HGSQYRGSRVESPVSYIGEDDDNNDYDFNE |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 24 .74 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 24 .45 RPM |
| SRP007412_heart | 0 .12 RPM | 14 .88 RPM |
| SRP007412_kidney | 0 .04 RPM | 15 .13 RPM |
| SRP007412_liver | 0 .00 RPM | 12 .32 RPM |
| SRP007412_testis | 0 .05 RPM | 27 .49 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_338 was not detected
No EST(s) were mapped for retro_mmus_338 retrocopy.
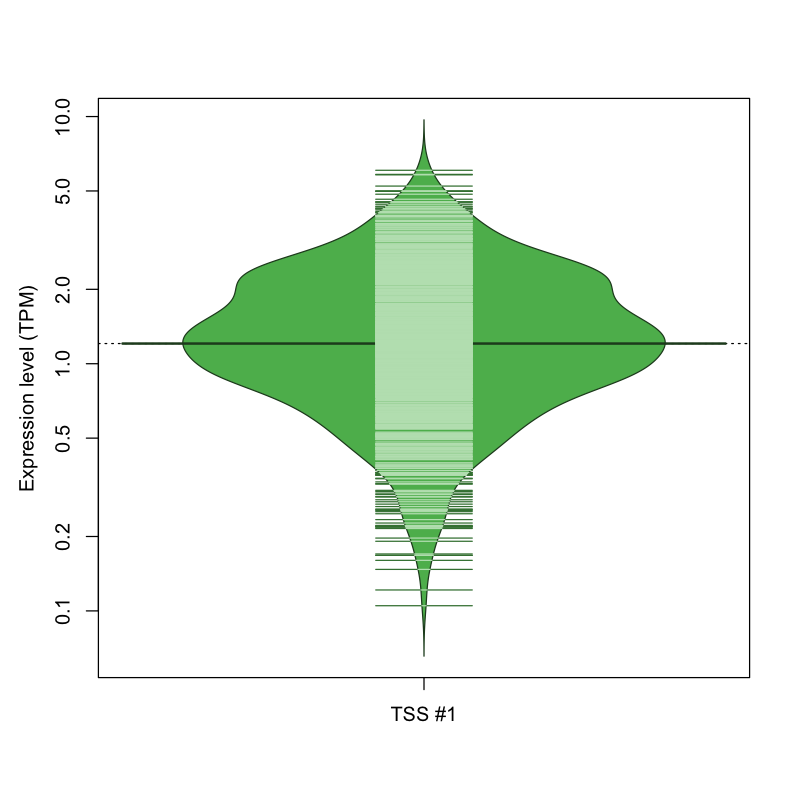
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_76595 | 189 libraries | 327 libraries | 551 libraries | 5 libraries | 0 libraries |
The graphical summary, for retro_mmus_338 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_338 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_338 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 28 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Choloepus hoffmanni | ENSCHOG00000004467 | 8 retrocopies | |
| Callithrix jacchus | ENSCJAG00000006950 | 2 retrocopies | |
| Homo sapiens | ENSG00000198176 | 4 retrocopies | |
| Gorilla gorilla | ENSGGOG00000004908 | 2 retrocopies | |
| Loxodonta africana | ENSLAFG00000007927 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000012165 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000007423 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000038482 | 2 retrocopies |
retro_mmus_3243, retro_mmus_338 ,
|
| Nomascus leucogenys | ENSNLEG00000000849 | 3 retrocopies | |
| Otolemur garnettii | ENSOGAG00000003532 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000025889 | 3 retrocopies |