RetrogeneDB ID: | retro_mmus_439 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:49399405..49400729(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Hspd1 | ||
| Ensembl ID: | ENSMUSG00000025980 | ||
| Aliases: | Hspd1, 60kDa, Hsp60 | ||
| Description: | heat shock protein 1 (chaperonin) [Source:MGI Symbol;Acc:MGI:96242] |

Retrocopy-Parental alignment summary:
>retro_mmus_439
ATGCCTCATCTATTCACAGTCCTTTGCCAGATGAGACCAGTGTCCCCAGCACTGGCTCCTCATCTCACTTGGGCCTATGC
CAAAGATATAAAATTTGGTGCGGATGCTTGAGCCTTAATGCTTCAAGGTATAGATCTTTTAGCTGATGCTATAGCTGTTA
CAGTGGGGTCAAAGCAAAGAACAGTGCTTATTGAATGGAGTTGGGGAAGTCCCAAATTAATAAAAGATAGGGTCACTGGC
CCAAAGTCAATTTCATAAAGGATAAATACAAAAATATCGGACATAAACTTGTTCAGGTTGTTGCCAATAACACAAACGAA
GAGGCTGGGGATGGTATTACCACTACCACTGTTCTGGCATGGTCTATTGCCAAGGAGGACTTTGAGAAGTGCAGCAAAGG
GCCATGAGAAATCAAAAGAAATGTGATGTTGGGTGTTAATGCTGTAATTACTGAACTTAAGAAACAATATAAACCTGTGA
CATCCCCTGATGAAATTGCTCAGGTTGCTACAATTTCTGGAAATAGAGACAAAGACATTTAAAATATCATTTCTGATAGA
ATGGAAAAGGTTGAAGAAAGGGTGTCATCACAGTGAAGGTTGGAAAAATCCTGAATGATGAAGTAGAAATTATTGAAGGC
ATGAAGTTTGATAGAGGATATATTTCTCTAGAATTTTATTAACACATCATAAGGTCAAAAAGTGTGAATTCCAAGATGCC
TATGTTTTTGTTGAGTGAAATGAAATTTTCTAGTGTCCAGTCCATTGTATCTGCTCTTAAAATTGTTAATGCTCATCAGA
TGCCCTTGGTAATAATTGCTGAAGATATTGATAGAGTAGCTCTAAGATCACTGATTTTGAACAAGCTAAAAGTTTGTCTT
CAGGTTGGAACAGTCAAAGCTCCAGGGTTTGGTGACAATCAGAAAAAACAGTTTAAAGATAATAGCTAATGCTACTAGTG
GTACAATGTTTTGAGAAGAGGGATTGAATCAAAACCTTGGAGATGTTCACGCTCATGTCATAGGAAAACTTGGAGAGGTC
CTTTTCACCAAAGAAGATGCCATGCTTTTGAAAGGAAAAGGTGACAAGGTTCACATTGAAAAACATATTCAAGAAATCAC
TGAACAGCTAGACATCACAACTGGTCAATATAAAAAGGAAAACTGAACAAGGGACTTGCTAAACTTTCAGATGGAGTAGG
TGTTTTGAAGGTTAGAGGAACAATTGATGTTGAAATGAAGGAGAAGACAGTTACAGATGCTCTCAGTGCTGCAAGAGCAG
CTGTTGAAGAATGCATTGTTCTTGGAGAAGGCTCTGCTTTGTTG
ATGCCTCATCTATTCACAGTCCTTTGCCAGATGAGACCAGTGTCCCCAGCACTGGCTCCTCATCTCACTTGGGCCTATGC
CAAAGATATAAAATTTGGTGCGGATGCTTGAGCCTTAATGCTTCAAGGTATAGATCTTTTAGCTGATGCTATAGCTGTTA
CAGTGGGGTCAAAGCAAAGAACAGTGCTTATTGAATGGAGTTGGGGAAGTCCCAAATTAATAAAAGATAGGGTCACTGGC
CCAAAGTCAATTTCATAAAGGATAAATACAAAAATATCGGACATAAACTTGTTCAGGTTGTTGCCAATAACACAAACGAA
GAGGCTGGGGATGGTATTACCACTACCACTGTTCTGGCATGGTCTATTGCCAAGGAGGACTTTGAGAAGTGCAGCAAAGG
GCCATGAGAAATCAAAAGAAATGTGATGTTGGGTGTTAATGCTGTAATTACTGAACTTAAGAAACAATATAAACCTGTGA
CATCCCCTGATGAAATTGCTCAGGTTGCTACAATTTCTGGAAATAGAGACAAAGACATTTAAAATATCATTTCTGATAGA
ATGGAAAAGGTTGAAGAAAGGGTGTCATCACAGTGAAGGTTGGAAAAATCCTGAATGATGAAGTAGAAATTATTGAAGGC
ATGAAGTTTGATAGAGGATATATTTCTCTAGAATTTTATTAACACATCATAAGGTCAAAAAGTGTGAATTCCAAGATGCC
TATGTTTTTGTTGAGTGAAATGAAATTTTCTAGTGTCCAGTCCATTGTATCTGCTCTTAAAATTGTTAATGCTCATCAGA
TGCCCTTGGTAATAATTGCTGAAGATATTGATAGAGTAGCTCTAAGATCACTGATTTTGAACAAGCTAAAAGTTTGTCTT
CAGGTTGGAACAGTCAAAGCTCCAGGGTTTGGTGACAATCAGAAAAAACAGTTTAAAGATAATAGCTAATGCTACTAGTG
GTACAATGTTTTGAGAAGAGGGATTGAATCAAAACCTTGGAGATGTTCACGCTCATGTCATAGGAAAACTTGGAGAGGTC
CTTTTCACCAAAGAAGATGCCATGCTTTTGAAAGGAAAAGGTGACAAGGTTCACATTGAAAAACATATTCAAGAAATCAC
TGAACAGCTAGACATCACAACTGGTCAATATAAAAAGGAAAACTGAACAAGGGACTTGCTAAACTTTCAGATGGAGTAGG
TGTTTTGAAGGTTAGAGGAACAATTGATGTTGAAATGAAGGAGAAGACAGTTACAGATGCTCTCAGTGCTGCAAGAGCAG
CTGTTGAAGAATGCATTGTTCTTGGAGAAGGCTCTGCTTTGTTG
ORF - retro_mmus_439 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 75. % |
| Parental protein coverage: | 77.66 % |
| Number of stop codons detected: | 5 |
| Number of frameshifts detected | 7 |
Retrocopy - Parental Gene Alignment:
| Parental | MLRLPTVLRQMRPVSRALAPHLTRAYAKDVKFGADARALMLQGVDLLADAVAVTMGPKGRTVIIEQSWGS |
| M..L.TVL.QMRPVS.ALAPHLT.AYAKD.KFGADA.ALMLQG.DLLADA.AVT.G.K.RTV.IE.SWGS | |
| Retrocopy | MPHLFTVLCQMRPVSPALAPHLTWAYAKDIKFGADA*ALMLQGIDLLADAIAVTVGSKQRTVLIEWSWGS |
| Parental | PKVTKDGVTVAKSID-LKDKYKNIGAKLVQDVANNTNEEAGDGTTTATVLARSIAKEGFEKISKGANPVE |
| PK..KD.VT..KSI...KDKYKNIG.KLVQ.VANNTNEEAGDG.TT.TVLA.SIAKE.FEK.SKG..P.E | |
| Retrocopy | PKLIKDRVTGPKSIS<IKDKYKNIGHKLVQVVANNTNEEAGDGITTTTVLAWSIAKEDFEKCSKG--P*E |
| Parental | IRRGVMLAVDAVIAELKKQSKPVTTPEEIAQVATISANGDKDIGNIISDAMKKVG-RKGVITVKDGKTLN |
| I.R.VML.V.AVI.ELKKQ.KPVT.P.EIAQVATIS.N.DKDI.NIISD.M.KV..RKGVITVK.GK.LN | |
| Retrocopy | IKRNVMLGVNAVITELKKQYKPVTSPDEIAQVATISGNRDKDI*NIISDRMEKVE<RKGVITVKVGKILN |
| Parental | DELEIIEGMKFDRGYISP-YFINTSKGQ-KCEFQDAYV-LLSEKKISSVQSIVPALEIANAHRKPLVIIA |
| DE.EIIEGMKFDRGYIS...FINTS.GQ.KCEFQDAYV.LLSE.K.SSVQSIV.AL.I.NAH..PLVIIA | |
| Retrocopy | DEVEIIEGMKFDRGYISL>NFINTS*GQ>KCEFQDAYV>LLSEMKFSSVQSIVSALKIVNAHQMPLVIIA |
| Parental | EDVDGEALSTLVLNRLKVGLQVVAVKAPGFGDN-RKNQLKDMAIATGGAVFGEEGLNLNLEDVQAHDLGK |
| ED.D..AL..L.LN.LKV.LQV..VKAPGFGDN.RKN.LK..A.AT.G..F.EEGLN.NL.DV.AH..GK | |
| Retrocopy | EDIDRVALRSLILNKLKVCLQVGTVKAPGFGDN>RKNSLKIIANATSGTMF*EEGLNQNLGDVHAHVIGK |
| Parental | VGEVIVTKDDAMLLKGKGDKAHIEKRIQEITEQLDITTSEYEKE-KLNERLAKLSDGVAVLKVGGTSDVE |
| .GEV..TK.DAMLLKGKGDK.HIEK.IQEITEQLDITT..Y.KE.KLN..LAKLSDGV.VLKV.GT.DVE | |
| Retrocopy | LGEVLFTKEDAMLLKGKGDKVHIEKHIQEITEQLDITTGQYKKE<KLNKGLAKLSDGVGVLKVRGTIDVE |
| Parental | VNEKKDRVTDALNATRAAVEEGIVLGGGCALL |
| ..EK...VTDAL.A.RAAVEE.IVLG.G.ALL | |
| Retrocopy | MKEK--TVTDALSAARAAVEECIVLGEGSALL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 93 .17 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 79 .33 RPM |
| SRP007412_heart | 0 .00 RPM | 210 .75 RPM |
| SRP007412_kidney | 0 .00 RPM | 298 .39 RPM |
| SRP007412_liver | 0 .00 RPM | 313 .50 RPM |
| SRP007412_testis | 0 .00 RPM | 22 .73 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_439 was not detected
No EST(s) were mapped for retro_mmus_439 retrocopy.
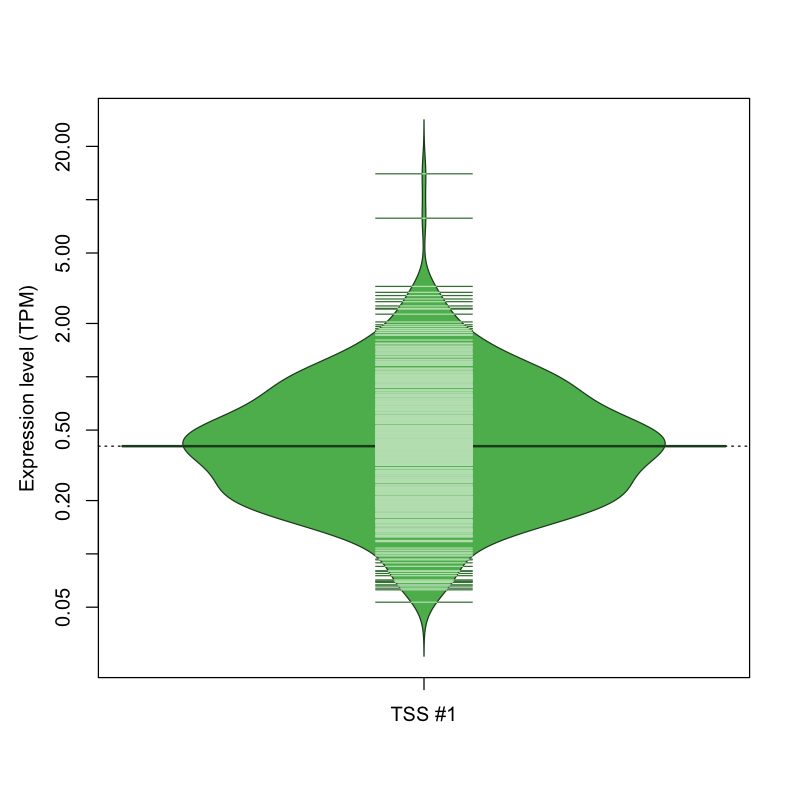
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_74689 | 576 libraries | 428 libraries | 66 libraries | 1 library | 1 library |
The graphical summary, for retro_mmus_439 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_439 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_439 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 18 parental genes, and 74 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000013770 | 1 retrocopy | |
| Canis familiaris | ENSCAFG00000010865 | 4 retrocopies | |
| Callithrix jacchus | ENSCJAG00000003990 | 6 retrocopies | |
| Echinops telfairi | ENSETEG00000009184 | 4 retrocopies | |
| Homo sapiens | ENSG00000144381 | 7 retrocopies | |
| Gorilla gorilla | ENSGGOG00000008267 | 6 retrocopies | |
| Loxodonta africana | ENSLAFG00000000053 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000011987 | 2 retrocopies | |
| Myotis lucifugus | ENSMLUG00000000871 | 4 retrocopies | |
| Macaca mulatta | ENSMMUG00000010489 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000012289 | 3 retrocopies | |
| Mustela putorius furo | ENSMPUG00000007818 | 3 retrocopies | |
| Mus musculus | ENSMUSG00000025980 | 5 retrocopies | |
| Nomascus leucogenys | ENSNLEG00000006874 | 7 retrocopies | |
| Otolemur garnettii | ENSOGAG00000034053 | 5 retrocopies | |
| Pongo abelii | ENSPPYG00000013046 | 5 retrocopies | |
| Rattus norvegicus | ENSRNOG00000014525 | 9 retrocopies | |
| Xenopus tropicalis | ENSXETG00000007509 | 1 retrocopy |