RetrogeneDB ID: | retro_hsap_2122 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 2:84517827..84518395(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000182814 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | FUNDC2 | ||
| Ensembl ID: | ENSG00000165775 | ||
| Aliases: | None | ||
| Description: | FUN14 domain containing 2 [Source:HGNC Symbol;Acc:24925] |

Retrocopy-Parental alignment summary:
>retro_hsap_2122
ATGGAAACATCTGCCCCACGTGCAGGAAGCCGGGTGGTGGCCACGACAGCGCGCCACTCCGCAGCGTAACCTCGCAGATC
CCCTGCGTGTGTCCTCGCAAGACTAGCTCACCGAAATGGCCGCGTCCAGTCAAGGAAACTTTGAGGGAGATATTGAGTCA
GTGGACCTTGCAGAATTTGCTAAGCAGCAGCCGTGGTGGCGTAAGCTGTTTGGGCCAGAATCTGGACTCTCAGCCGAAAA
GTATAGCGTGGCAACCCACCTGTTCATTGGAGGTGTCACTGGATGGTGCACGGGTTTTATATTCCAGAAGGTTGGAAAGT
TGGCTGCAACAGCTGTGGGAGGTGGATTTTTTCTCCTTCAGCTTGCAAACCATTCTGGGTACATCAAAGTTGATTGGCAA
CGAGTGGAGAAGGACATGAAGAAAGCCAAAGAGCAGCTGAAGATCCCTAAGAGCACTCAGATACCTAACCAGGTCAGGAG
CAAAGCTGAGGAGGTGGTGTCATTTGTGAAGAAAAATGTTCTAGTGACTGGGGGATTTTTCGGAGGCTTTCTGCTTGGCA
TGGCATCC
ATGGAAACATCTGCCCCACGTGCAGGAAGCCGGGTGGTGGCCACGACAGCGCGCCACTCCGCAGCGTAACCTCGCAGATC
CCCTGCGTGTGTCCTCGCAAGACTAGCTCACCGAAATGGCCGCGTCCAGTCAAGGAAACTTTGAGGGAGATATTGAGTCA
GTGGACCTTGCAGAATTTGCTAAGCAGCAGCCGTGGTGGCGTAAGCTGTTTGGGCCAGAATCTGGACTCTCAGCCGAAAA
GTATAGCGTGGCAACCCACCTGTTCATTGGAGGTGTCACTGGATGGTGCACGGGTTTTATATTCCAGAAGGTTGGAAAGT
TGGCTGCAACAGCTGTGGGAGGTGGATTTTTTCTCCTTCAGCTTGCAAACCATTCTGGGTACATCAAAGTTGATTGGCAA
CGAGTGGAGAAGGACATGAAGAAAGCCAAAGAGCAGCTGAAGATCCCTAAGAGCACTCAGATACCTAACCAGGTCAGGAG
CAAAGCTGAGGAGGTGGTGTCATTTGTGAAGAAAAATGTTCTAGTGACTGGGGGATTTTTCGGAGGCTTTCTGCTTGGCA
TGGCATCC
ORF - retro_hsap_2122 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 90.53 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | METSAPRAGSQVVATTARHSAA-YRADPLRVSSRDKLTEMAASSQGNFEGNFESLDLAEFAKKQPWWRKL |
| METSAPRAGS.VVATTARHSAA...ADPLRVSS.D.LTEMAASSQGNFEG..ES.DLAEFAK.QPWWRKL | |
| Retrocopy | METSAPRAGSRVVATTARHSAA>NLADPLRVSSQD*LTEMAASSQGNFEGDIESVDLAEFAKQQPWWRKL |
| Parental | FGQESGPSAEKYSVATQLFIGGVTGWCTGFIFQKVGKLAATAVGGGFFLLQLANHTGYIKVDWQRVEKDM |
| FG.ESG.SAEKYSVAT.LFIGGVTGWCTGFIFQKVGKLAATAVGGGFFLLQLANH.GYIKVDWQRVEKDM | |
| Retrocopy | FGPESGLSAEKYSVATHLFIGGVTGWCTGFIFQKVGKLAATAVGGGFFLLQLANHSGYIKVDWQRVEKDM |
| Parental | KKAKEQLKIRKSNQIPTEVRSKAEEVVSFVKKNVLVTGGFFGGFLLGMAS |
| KKAKEQLKI.KS.QIP..VRSKAEEVVSFVKKNVLVTGGFFGGFLLGMAS | |
| Retrocopy | KKAKEQLKIPKSTQIPNQVRSKAEEVVSFVKKNVLVTGGFFGGFLLGMAS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 44 .98 RPM |
| bodymap2_adrenal | 0 .00 RPM | 85 .17 RPM |
| bodymap2_brain | 0 .00 RPM | 43 .53 RPM |
| bodymap2_breast | 0 .00 RPM | 63 .89 RPM |
| bodymap2_colon | 0 .00 RPM | 87 .75 RPM |
| bodymap2_heart | 0 .00 RPM | 123 .45 RPM |
| bodymap2_kidney | 0 .00 RPM | 72 .90 RPM |
| bodymap2_liver | 0 .00 RPM | 38 .55 RPM |
| bodymap2_lung | 0 .00 RPM | 66 .70 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 80 .84 RPM |
| bodymap2_ovary | 0 .00 RPM | 96 .92 RPM |
| bodymap2_prostate | 0 .00 RPM | 78 .20 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 57 .15 RPM |
| bodymap2_testis | 4 .77 RPM | 85 .27 RPM |
| bodymap2_thyroid | 0 .13 RPM | 99 .22 RPM |
| bodymap2_white_blood_cells | 0 .49 RPM | 38 .12 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_2122 was not detected
1 EST(s) were mapped to retro_hsap_2122 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| DB079965 | 84517834 | 84518072 | 100 | 238 | 0 | 238 |
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_125593 | 1799 libraries | 23 libraries | 4 libraries | 1 library | 2 libraries |
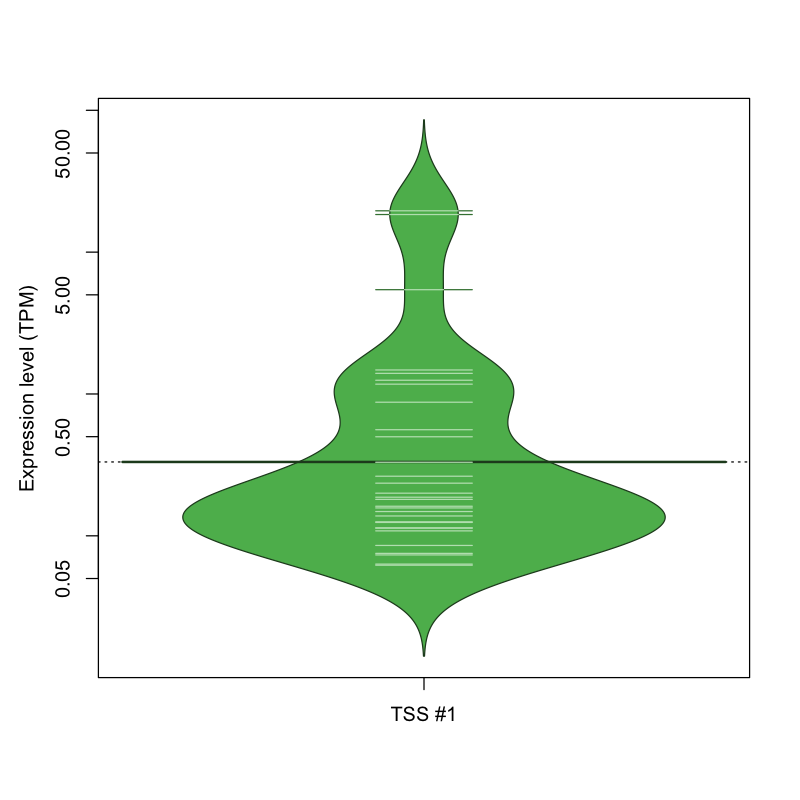
The graphical summary, for retro_hsap_2122 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_2122 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2122 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 20 parental genes, and 50 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .04 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .04 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .00 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .03 RPM |
| FIN_HG00375 | 0 .02 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .02 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .03 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .03 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .00 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .03 RPM |
| TSI_NA20765 | 0 .00 RPM |
| TSI_NA20771 | 0 .03 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .03 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .02 RPM |