RetrogeneDB ID: | retro_hsap_1563 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 15:44281261..44282382(-) | ||
| Located in intron of: | ENSG00000171877 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000185607 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | ACTB | ||
| Ensembl ID: | ENSG00000075624 | ||
| Aliases: | ACTB, BRWS1, PS1TP5BP1 | ||
| Description: | actin, beta [Source:HGNC Symbol;Acc:132] |

Retrocopy-Parental alignment summary:
>retro_hsap_1563
ATGGATGATGATATCACCATGCTTGTCGTCGACAATGGCTCTGGCATGTGCAAGGCCAGCTTTGCAGGTGACGATGCCCA
CCCCGGGCCATCTTCCCCTTCATTGTGGGGCGCCCCAGACACCAGGGCGTGACGGTGGGCATGGGTCAGAAGGACTCCTA
TGTGGGCAACGAGGTCCAGAGCAAGAGAGGCATCCTGACCCTGAAGTACCCCATCAAGCAGGACATTGTCACCACCTGGG
ACGACATGGAGAAGATCTGGCACCACACCTTCTACAACGAGCTGTGTGTGGCTGCCAAGGAGCACCCCATGCTGCTGACC
AAGGTCCCCCTGAGCCCCAAGGCCAACCACAAGAAGATGACCCAGATCATGTTGGAGATCTTCGACAGGCCAGCCATGTA
CGTGGCCATCCAGGCCGTGCTGTCCCTGTACACCTCTGGCCTACCACTGACATCGTGATGGACTACGATGACGGGGTCAC
CCACACTGTGCCCATCTATGAAGAGTATGCCCTCCCCCATGCCATCCTGCGTGTGTGCCTGGCTGGTCAGGACCTGACTG
ACTACCTCATGAAGGTCCTCACCTAGCACAGCTACATCTTCACCGCCACAGCCAAGCGGGAAATTGTGCATGACATCAAG
GAGAAGCTGTGCTATGTCACCCTGGACTTCGAGCAGGAGATGGCCATGGTGGCCTCCAGCTCCTCCCTGGAGATAGCTAC
AAGCTGCCCGACGGCCAGGTCATCATCATCGGCAAAAGCAGTTCTGCTGCCCTGAGGCGCTCTTCCAGCCTTCCTTCCTG
GGCATGGAATCCCGCGGCATCCATGAAACTACCTTCAACTCCATCATGAAGTGTGATGTGGACATCCTCAAAGACCCGTA
TGCCAACACAGTGCTATCTGGCGGCACCACCAGGTACCCTGGCATCGCCGATAGGATGCAGAAGATCACCACCCTGGCGC
CCAGCACAATGAAGATCAAGATCACTGCTCCTCCCAAGCGCAAGTACTCCGTGTGGATCGGCAGCTCCATCCTGGCCTGG
CTGTCCACCGTCCAGCAGATGTGGATCAGCAAGCAGGAATATGATGAGTCAGGCACCTCCATCATCCACCGCAGATGCTT
C
ATGGATGATGATATCACCATGCTTGTCGTCGACAATGGCTCTGGCATGTGCAAGGCCAGCTTTGCAGGTGACGATGCCCA
CCCCGGGCCATCTTCCCCTTCATTGTGGGGCGCCCCAGACACCAGGGCGTGACGGTGGGCATGGGTCAGAAGGACTCCTA
TGTGGGCAACGAGGTCCAGAGCAAGAGAGGCATCCTGACCCTGAAGTACCCCATCAAGCAGGACATTGTCACCACCTGGG
ACGACATGGAGAAGATCTGGCACCACACCTTCTACAACGAGCTGTGTGTGGCTGCCAAGGAGCACCCCATGCTGCTGACC
AAGGTCCCCCTGAGCCCCAAGGCCAACCACAAGAAGATGACCCAGATCATGTTGGAGATCTTCGACAGGCCAGCCATGTA
CGTGGCCATCCAGGCCGTGCTGTCCCTGTACACCTCTGGCCTACCACTGACATCGTGATGGACTACGATGACGGGGTCAC
CCACACTGTGCCCATCTATGAAGAGTATGCCCTCCCCCATGCCATCCTGCGTGTGTGCCTGGCTGGTCAGGACCTGACTG
ACTACCTCATGAAGGTCCTCACCTAGCACAGCTACATCTTCACCGCCACAGCCAAGCGGGAAATTGTGCATGACATCAAG
GAGAAGCTGTGCTATGTCACCCTGGACTTCGAGCAGGAGATGGCCATGGTGGCCTCCAGCTCCTCCCTGGAGATAGCTAC
AAGCTGCCCGACGGCCAGGTCATCATCATCGGCAAAAGCAGTTCTGCTGCCCTGAGGCGCTCTTCCAGCCTTCCTTCCTG
GGCATGGAATCCCGCGGCATCCATGAAACTACCTTCAACTCCATCATGAAGTGTGATGTGGACATCCTCAAAGACCCGTA
TGCCAACACAGTGCTATCTGGCGGCACCACCAGGTACCCTGGCATCGCCGATAGGATGCAGAAGATCACCACCCTGGCGC
CCAGCACAATGAAGATCAAGATCACTGCTCCTCCCAAGCGCAAGTACTCCGTGTGGATCGGCAGCTCCATCCTGGCCTGG
CTGTCCACCGTCCAGCAGATGTGGATCAGCAAGCAGGAATATGATGAGTCAGGCACCTCCATCATCCACCGCAGATGCTT
C
ORF - retro_hsap_1563 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 80.79 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 4 |
Retrocopy - Parental Gene Alignment:
| Parental | MDDDIAALVVDNGSGMCKAGFAGDDA-PRAVFPS-IVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLK |
| MDDDI..LVVDNGSGMCKA.FAGDDA.P....PS.IVGRPRHQGV.VGMGQKDSYVG.E.QSKRGILTLK | |
| Retrocopy | MDDDITMLVVDNGSGMCKASFAGDDAHPGPSSPS<IVGRPRHQGVTVGMGQKDSYVGNEVQSKRGILTLK |
| Parental | YPIEHGIVTNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMTQIMFETFNTPAMYVAIQA |
| YPI...IVT.WDDMEKIWHHTFYNEL.VA..EHP.LLT..PL.PKAN..KMTQIM.E.F..PAMYVAIQA | |
| Retrocopy | YPIKQDIVTTWDDMEKIWHHTFYNELCVAAKEHPMLLTKVPLSPKANHKKMTQIMLEIFDRPAMYVAIQA |
| Parental | VLSLYASG-RTTGIVMDSGDGVTHTVPIYEGYALPHAILRLDLAGRDLTDYLMKILTERGYSFTTTAERE |
| VLSLY.SG..TT.IVMD..DGVTHTVPIYE.YALPHAILR..LAG.DLTDYLMK.LT...Y.FT.TA.RE | |
| Retrocopy | VLSLYTSG<PTTDIVMDYDDGVTHTVPIYEEYALPHAILRVCLAGQDLTDYLMKVLT*HSYIFTATAKRE |
| Parental | IVRDIKEKLCYVALDFEQEMATAASSSSLE-KSYELPDGQVITIG-NERFRCPEALFQPSFLGMESCGIH |
| IV.DIKEKLCYV.LDFEQEMA..ASSSSLE..SY.LPDGQVI.IG....F.CPEALFQPSFLGMES.GIH | |
| Retrocopy | IVHDIKEKLCYVTLDFEQEMAMVASSSSLE<DSYKLPDGQVIIIG<QKQFCCPEALFQPSFLGMESRGIH |
| Parental | ETTFNSIMKCDVDIRKDLYANTVLSGGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSI |
| ETTFNSIMKCDVDI.KD.YANTVLSGGTT.YPGIADRMQK.IT.LAPSTMKIKI.APP.RKYSVWIG.SI | |
| Retrocopy | ETTFNSIMKCDVDILKDPYANTVLSGGTTRYPGIADRMQK-ITTLAPSTMKIKITAPPKRKYSVWIGSSI |
| Parental | LASLSTFQQMWISKQEYDESGPSIVHRKCF |
| LA.LST.QQMWISKQEYDESG.SI.HR.CF | |
| Retrocopy | LAWLSTVQQMWISKQEYDESGTSIIHRRCF |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .02 RPM | 5336 .71 RPM |
| bodymap2_adrenal | 0 .02 RPM | 5105 .37 RPM |
| bodymap2_brain | 0 .16 RPM | 2354 .74 RPM |
| bodymap2_breast | 0 .02 RPM | 3342 .68 RPM |
| bodymap2_colon | 0 .02 RPM | 6846 .79 RPM |
| bodymap2_heart | 0 .02 RPM | 595 .89 RPM |
| bodymap2_kidney | 0 .10 RPM | 2141 .79 RPM |
| bodymap2_liver | 0 .00 RPM | 1498 .34 RPM |
| bodymap2_lung | 0 .00 RPM | 6019 .68 RPM |
| bodymap2_lymph_node | 0 .02 RPM | 3028 .73 RPM |
| bodymap2_ovary | 0 .12 RPM | 4495 .81 RPM |
| bodymap2_prostate | 0 .07 RPM | 6984 .69 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 729 .01 RPM |
| bodymap2_testis | 0 .19 RPM | 2997 .25 RPM |
| bodymap2_thyroid | 0 .23 RPM | 1886 .18 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 9289 .16 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_1563 was not detected
1 EST(s) were mapped to retro_hsap_1563 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| CN273804 | 44281349 | 44281824 | 100 | 475 | 0 | 475 |
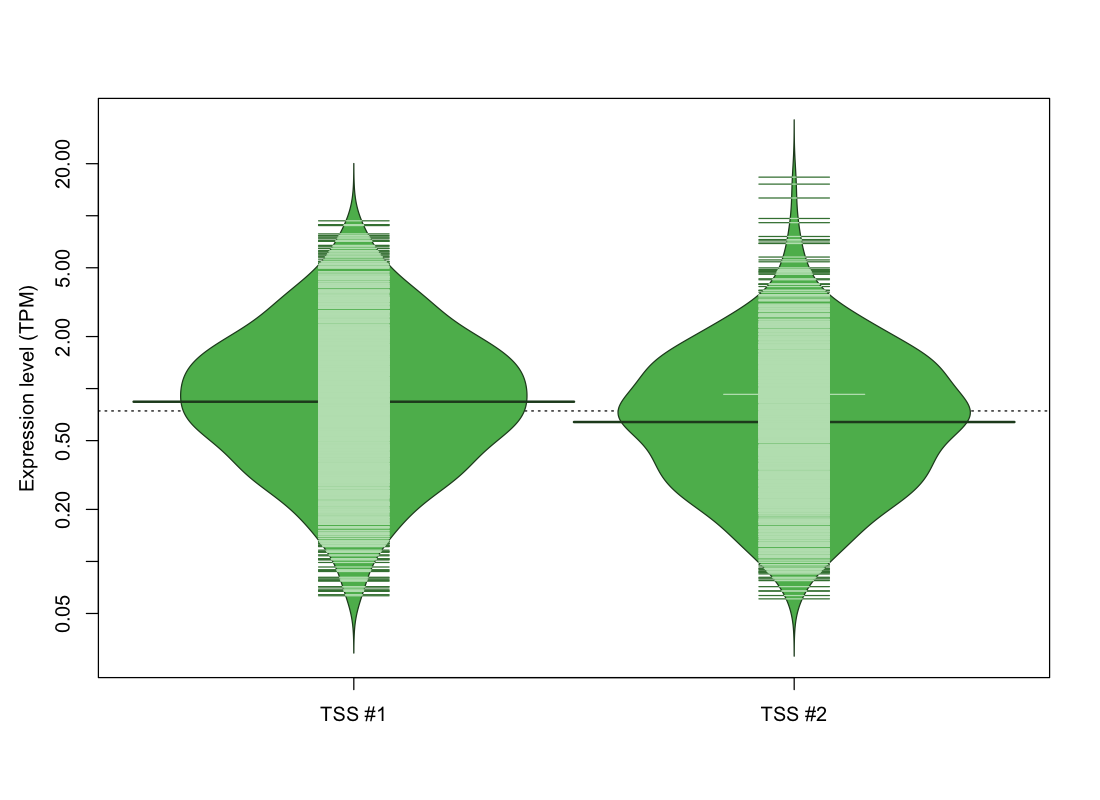
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_43836 | 599 libraries | 690 libraries | 504 libraries | 36 libraries | 0 libraries |
| TSS #2 | TSS_43837 | 807 libraries | 686 libraries | 323 libraries | 10 libraries | 3 libraries |
The graphical summary, for retro_hsap_1563 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_1563 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_1563 has 3 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_1051 |
| Gorilla gorilla | retro_ggor_1188 |
| Pongo abelii | retro_pabe_1292 |
Parental genes homology:
Parental genes homology involve 22 parental genes, and 99 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .09 RPM |
| CEU_NA11843 | 0 .11 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .04 RPM |
| CEU_NA12400 | 0 .11 RPM |
| CEU_NA12751 | 0 .12 RPM |
| CEU_NA12760 | 0 .13 RPM |
| CEU_NA12827 | 0 .05 RPM |
| CEU_NA12872 | 0 .11 RPM |
| CEU_NA12873 | 0 .03 RPM |
| FIN_HG00183 | 0 .03 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .06 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .04 RPM |
| FIN_HG00349 | 0 .06 RPM |
| FIN_HG00375 | 0 .02 RPM |
| FIN_HG00377 | 0 .03 RPM |
| FIN_HG00378 | 0 .08 RPM |
| GBR_HG00099 | 0 .06 RPM |
| GBR_HG00111 | 0 .04 RPM |
| GBR_HG00114 | 0 .05 RPM |
| GBR_HG00119 | 0 .02 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .07 RPM |
| GBR_HG00134 | 0 .02 RPM |
| GBR_HG00137 | 0 .03 RPM |
| GBR_HG00142 | 0 .08 RPM |
| GBR_HG00143 | 0 .03 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .03 RPM |
| TSI_NA20532 | 0 .14 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .12 RPM |
| TSI_NA20765 | 0 .02 RPM |
| TSI_NA20771 | 0 .11 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .03 RPM |
| YRI_NA18870 | 0 .03 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .05 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .06 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .02 RPM |
| YRI_NA19223 | 0 .04 RPM |