RetrogeneDB ID: | retro_hsap_2568 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 22:17075975..17076378(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000240122 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | FABP5 | ||
| Ensembl ID: | ENSG00000164687 | ||
| Aliases: | FABP5, E-FABP, EFABP, KFABP, PA-FABP, PAFABP | ||
| Description: | fatty acid binding protein 5 (psoriasis-associated) [Source:HGNC Symbol;Acc:3560] |

Retrocopy-Parental alignment summary:
>retro_hsap_2568
ATGGCCACAGTTCAGCAGCTGGAAGGAAGATGGCGCCTGGTGGACAGCGAAGGCTTTGATGAATACATGAAGGAGCTAGG
AGGAAATAGCTTTGCAAAAAATGGGCGCAATGGCCAAGCCAGATTGTATCATCACTTGTGATGGCAAAAACCTCACCATA
AAAACTGAGAGCACTTTGAAAACAACACAGTTTTCTTGTACCCTGGGAGAGAAGTTTGAAGAAACCACAGCTGATGGCAG
AAAAACTCAGACTGTGTGCAACTTTACAGATGGTGCATTGGTTCAGCATCAGGAGTGGGATGGGAAGGAAAGCACAATAA
CAAGAACATTGAAAGATGGGAAATTAGTGGTGGACTGTGTCATGAACCATGTCGCCTGTACTCGGATCTATGAAAAAGTA
CAA
ATGGCCACAGTTCAGCAGCTGGAAGGAAGATGGCGCCTGGTGGACAGCGAAGGCTTTGATGAATACATGAAGGAGCTAGG
AGGAAATAGCTTTGCAAAAAATGGGCGCAATGGCCAAGCCAGATTGTATCATCACTTGTGATGGCAAAAACCTCACCATA
AAAACTGAGAGCACTTTGAAAACAACACAGTTTTCTTGTACCCTGGGAGAGAAGTTTGAAGAAACCACAGCTGATGGCAG
AAAAACTCAGACTGTGTGCAACTTTACAGATGGTGCATTGGTTCAGCATCAGGAGTGGGATGGGAAGGAAAGCACAATAA
CAAGAACATTGAAAGATGGGAAATTAGTGGTGGACTGTGTCATGAACCATGTCGCCTGTACTCGGATCTATGAAAAAGTA
CAA
ORF - retro_hsap_2568 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 91.18 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MATVQQLEGRWRLVDSKGFDEYMKELGVGIALRK-MGAMAKPDCIITCDGKNLTIKTESTLKTTQFSCTL |
| MATVQQLEGRWRLVDS.GFDEYMKELG.G....K.MGAMAKPDCIITCDGKNLTIKTESTLKTTQFSCTL | |
| Retrocopy | MATVQQLEGRWRLVDSEGFDEYMKELG-GNSFAK>MGAMAKPDCIITCDGKNLTIKTESTLKTTQFSCTL |
| Parental | GEKFEETTADGRKTQTVCNFTDGALVQHQEWDGKESTITRKLKDGKLVVECVMNNVTCTRIYEKVE |
| GEKFEETTADGRKTQTVCNFTDGALVQHQEWDGKESTITR.LKDGKLVV.CVMN.V.CTRIYEKV. | |
| Retrocopy | GEKFEETTADGRKTQTVCNFTDGALVQHQEWDGKESTITRTLKDGKLVVDCVMNHVACTRIYEKVQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 19 .10 RPM |
| bodymap2_adrenal | 0 .02 RPM | 30 .15 RPM |
| bodymap2_brain | 0 .00 RPM | 20 .77 RPM |
| bodymap2_breast | 0 .02 RPM | 44 .65 RPM |
| bodymap2_colon | 0 .00 RPM | 21 .44 RPM |
| bodymap2_heart | 0 .00 RPM | 14 .62 RPM |
| bodymap2_kidney | 0 .02 RPM | 2 .48 RPM |
| bodymap2_liver | 0 .00 RPM | 0 .42 RPM |
| bodymap2_lung | 0 .00 RPM | 14 .24 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 31 .58 RPM |
| bodymap2_ovary | 0 .02 RPM | 4 .59 RPM |
| bodymap2_prostate | 0 .00 RPM | 21 .62 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 3 .60 RPM |
| bodymap2_testis | 0 .00 RPM | 11 .88 RPM |
| bodymap2_thyroid | 0 .06 RPM | 7 .38 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 1 .16 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_2568 was not detected
No EST(s) were mapped for retro_hsap_2568 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_106383 | 1339 libraries | 390 libraries | 97 libraries | 2 libraries | 1 library |
The graphical summary, for retro_hsap_2568 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
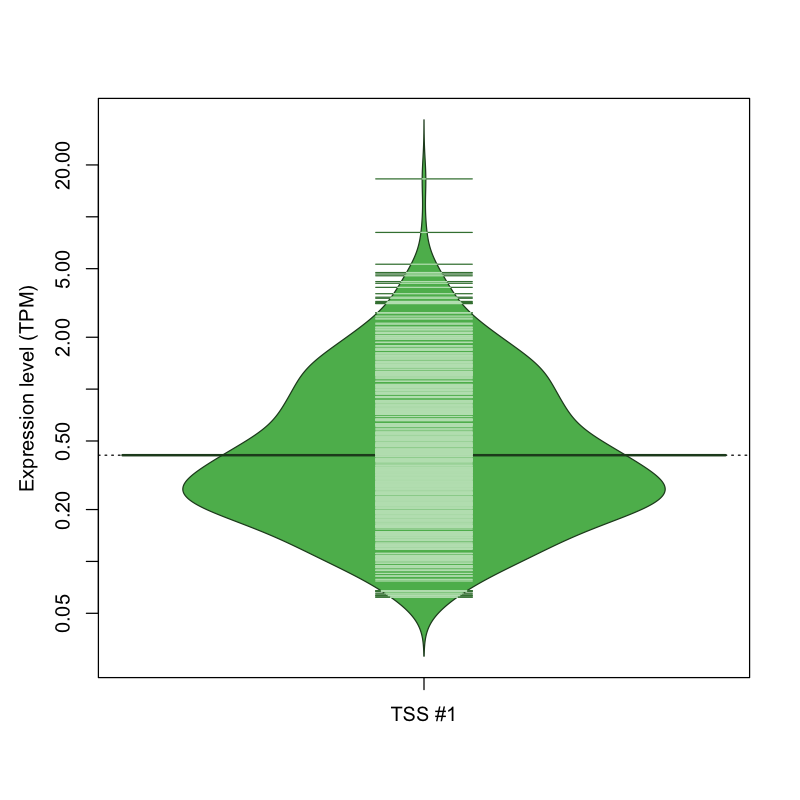
retro_hsap_2568 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2568 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_1498 |
Parental genes homology:
Parental genes homology involve 29 parental genes, and 108 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .06 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .00 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .04 RPM |
| CEU_NA12827 | 0 .05 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .03 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .02 RPM |
| FIN_HG00349 | 0 .03 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .00 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .03 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .00 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .02 RPM |
| YRI_NA19223 | 0 .02 RPM |