RetrogeneDB ID: | retro_mmus_135 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 10:62071122..62072361(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000046687 | |
| Aliases: | Gm5424, EG432466 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ass1 | ||
| Ensembl ID: | ENSMUSG00000076441 | ||
| Aliases: | Ass1, AA408052, ASS, Ass-1, fold | ||
| Description: | argininosuccinate synthetase 1 [Source:MGI Symbol;Acc:MGI:88090] |

Retrocopy-Parental alignment summary:
>retro_mmus_135
ATGTCCAGCAAGGGCTCTGTGGTTCTGGCCTACAGTGGTGGCCTGGACACCTCCTGCATCCTCGTGTGGCTGAAGGAACA
AGGCTATGATGTCATCGCCTACCTGGCCAACATTGGCCAGAAGGAAGACTTTGAGGAAGCCAGGAAGAAGGCGCTGAAGC
TTGGGGCCAAAAAGGTGTTCATTGAGGATGTGAGCAAGGAATTTGTGGAAGAGTTCATCTGGCCTGCTGTCCAGTCCAGT
GCACTCTACGAGGACCGCTATCTCCTGGGCACCTCTCTCGCCAGGCCTTGCATAGCTCGCAGACAGGTGGAGATTGCCCA
GCGTGAAGGGGCCAAGTATGTGTCTCACGGCGCCACGGGAAAGGGGAATGACCAGGTCCGCTTTGAGCTCACCTGCTACT
CACTGGCACCCCAGATTAAGGTCATCGCCCCCTGGAGGATGCCTGAGTTTTACAACCGGTTCAAGGGCCGAAATGATCTG
ATGGAGTATGCAAAGCAACACGGAATCCCCATCCCTGTCTCCCCCAAGAGCCCCTGGAGTATGGATGAAAACCTCATGCA
CATCAGCTATGAGGCTGGGATCCTGGAAAACCCCAAGAATCAAGCACCTCCGGGTCTCTACACAAAAACTCAGGACCCTG
CCAAAGCACCCAACACCCCAGATGTCCTTGAGATAGAATTCAAAAAAGGGGTCCCTGTGAAGGTGACCAACATCAAAGAT
GGCACAACCCGCACCACATCCCTGGAACTCTTCATGTACCTGAACGAAGTTGCGGGCAAGCACGGAGTGGGTCGCATTGA
CATCGTGGAGAACCGCTTCATTGGAATGAAGTCCCGAGGTATCTACGAGACCCCAGCAGGGACCATCCTTTACCACGCTC
ATTTAGACATAGAGGCCTTCACGATGGATCGGGAAGTACGCAAAATCAAGCAGGGCCTGGGCCTCAAATTCGCAGAGCTC
GTATACACAGGTTTCTGGCACAGCCCTGAATGTGAATTTGTTCGCCACTGCATCCAGAAGTCCCAGGAGCGGGTAGAAGG
GAAGGTGCAGGTGTCTGTCTTCAAGGGCCAAGTGTACATCCTCGGTCGGGAGTCTCCACTTTCACTCTACAATGAAGAGC
TGGTGAGCATGAACGTGCAGGGTGACTATGAGCCCATCGACGCCACTGGCTTCATCAATATCAACTCGCTCAGGCTGAAG
GAGTACCATCGCCTTCAGAGCAAGGTCACTGCCAAATAG
ATGTCCAGCAAGGGCTCTGTGGTTCTGGCCTACAGTGGTGGCCTGGACACCTCCTGCATCCTCGTGTGGCTGAAGGAACA
AGGCTATGATGTCATCGCCTACCTGGCCAACATTGGCCAGAAGGAAGACTTTGAGGAAGCCAGGAAGAAGGCGCTGAAGC
TTGGGGCCAAAAAGGTGTTCATTGAGGATGTGAGCAAGGAATTTGTGGAAGAGTTCATCTGGCCTGCTGTCCAGTCCAGT
GCACTCTACGAGGACCGCTATCTCCTGGGCACCTCTCTCGCCAGGCCTTGCATAGCTCGCAGACAGGTGGAGATTGCCCA
GCGTGAAGGGGCCAAGTATGTGTCTCACGGCGCCACGGGAAAGGGGAATGACCAGGTCCGCTTTGAGCTCACCTGCTACT
CACTGGCACCCCAGATTAAGGTCATCGCCCCCTGGAGGATGCCTGAGTTTTACAACCGGTTCAAGGGCCGAAATGATCTG
ATGGAGTATGCAAAGCAACACGGAATCCCCATCCCTGTCTCCCCCAAGAGCCCCTGGAGTATGGATGAAAACCTCATGCA
CATCAGCTATGAGGCTGGGATCCTGGAAAACCCCAAGAATCAAGCACCTCCGGGTCTCTACACAAAAACTCAGGACCCTG
CCAAAGCACCCAACACCCCAGATGTCCTTGAGATAGAATTCAAAAAAGGGGTCCCTGTGAAGGTGACCAACATCAAAGAT
GGCACAACCCGCACCACATCCCTGGAACTCTTCATGTACCTGAACGAAGTTGCGGGCAAGCACGGAGTGGGTCGCATTGA
CATCGTGGAGAACCGCTTCATTGGAATGAAGTCCCGAGGTATCTACGAGACCCCAGCAGGGACCATCCTTTACCACGCTC
ATTTAGACATAGAGGCCTTCACGATGGATCGGGAAGTACGCAAAATCAAGCAGGGCCTGGGCCTCAAATTCGCAGAGCTC
GTATACACAGGTTTCTGGCACAGCCCTGAATGTGAATTTGTTCGCCACTGCATCCAGAAGTCCCAGGAGCGGGTAGAAGG
GAAGGTGCAGGTGTCTGTCTTCAAGGGCCAAGTGTACATCCTCGGTCGGGAGTCTCCACTTTCACTCTACAATGAAGAGC
TGGTGAGCATGAACGTGCAGGGTGACTATGAGCCCATCGACGCCACTGGCTTCATCAATATCAACTCGCTCAGGCTGAAG
GAGTACCATCGCCTTCAGAGCAAGGTCACTGCCAAATAG
ORF - retro_mmus_135 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 99.51 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSSKGSVVLAYSGGLDTSCILVWLKEQGYDVIAYLANIGQKEDFEEARKKALKLGAKKVFIEDVSKEFVE |
| MSSKGSVVLAYSGGLDTSCILVWLKEQGYDVIAYLANIGQKEDFEEARKKALKLGAKKVFIEDVSKEFVE | |
| Retrocopy | MSSKGSVVLAYSGGLDTSCILVWLKEQGYDVIAYLANIGQKEDFEEARKKALKLGAKKVFIEDVSKEFVE |
| Parental | EFIWPAVQSSALYEDRYLLGTSLARPCIARRQVEIAQREGAKYVSHGATGKGNDQVRFELTCYSLAPQIK |
| EFIWPAVQSSALYEDRYLLGTSLARPCIARRQVEIAQREGAKYVSHGATGKGNDQVRFELTCYSLAPQIK | |
| Retrocopy | EFIWPAVQSSALYEDRYLLGTSLARPCIARRQVEIAQREGAKYVSHGATGKGNDQVRFELTCYSLAPQIK |
| Parental | VIAPWRMPEFYNRFKGRNDLMEYAKQHGIPIPVTPKSPWSMDENLMHISYEAGILENPKNQAPPGLYTKT |
| VIAPWRMPEFYNRFKGRNDLMEYAKQHGIPIPV.PKSPWSMDENLMHISYEAGILENPKNQAPPGLYTKT | |
| Retrocopy | VIAPWRMPEFYNRFKGRNDLMEYAKQHGIPIPVSPKSPWSMDENLMHISYEAGILENPKNQAPPGLYTKT |
| Parental | QDPAKAPNSPDVLEIEFKKGVPVKVTNIKDGTTRTTSLELFMYLNEVAGKHGVGRIDIVENRFIGMKSRG |
| QDPAKAPN.PDVLEIEFKKGVPVKVTNIKDGTTRTTSLELFMYLNEVAGKHGVGRIDIVENRFIGMKSRG | |
| Retrocopy | QDPAKAPNTPDVLEIEFKKGVPVKVTNIKDGTTRTTSLELFMYLNEVAGKHGVGRIDIVENRFIGMKSRG |
| Parental | IYETPAGTILYHAHLDIEAFTMDREVRKIKQGLGLKFAELVYTGFWHSPECEFVRHCIQKSQERVEGKVQ |
| IYETPAGTILYHAHLDIEAFTMDREVRKIKQGLGLKFAELVYTGFWHSPECEFVRHCIQKSQERVEGKVQ | |
| Retrocopy | IYETPAGTILYHAHLDIEAFTMDREVRKIKQGLGLKFAELVYTGFWHSPECEFVRHCIQKSQERVEGKVQ |
| Parental | VSVFKGQVYILGRESPLSLYNEELVSMNVQGDYEPIDATGFININSLRLKEYHRLQSKVTAK |
| VSVFKGQVYILGRESPLSLYNEELVSMNVQGDYEPIDATGFININSLRLKEYHRLQSKVTAK | |
| Retrocopy | VSVFKGQVYILGRESPLSLYNEELVSMNVQGDYEPIDATGFININSLRLKEYHRLQSKVTAK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .14 RPM | 13 .29 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 7 .34 RPM |
| SRP007412_heart | 0 .00 RPM | 2 .12 RPM |
| SRP007412_kidney | 15 .05 RPM | 584 .63 RPM |
| SRP007412_liver | 21 .52 RPM | 1275 .99 RPM |
| SRP007412_testis | 0 .05 RPM | 13 .95 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_135 was not detected
No EST(s) were mapped for retro_mmus_135 retrocopy.
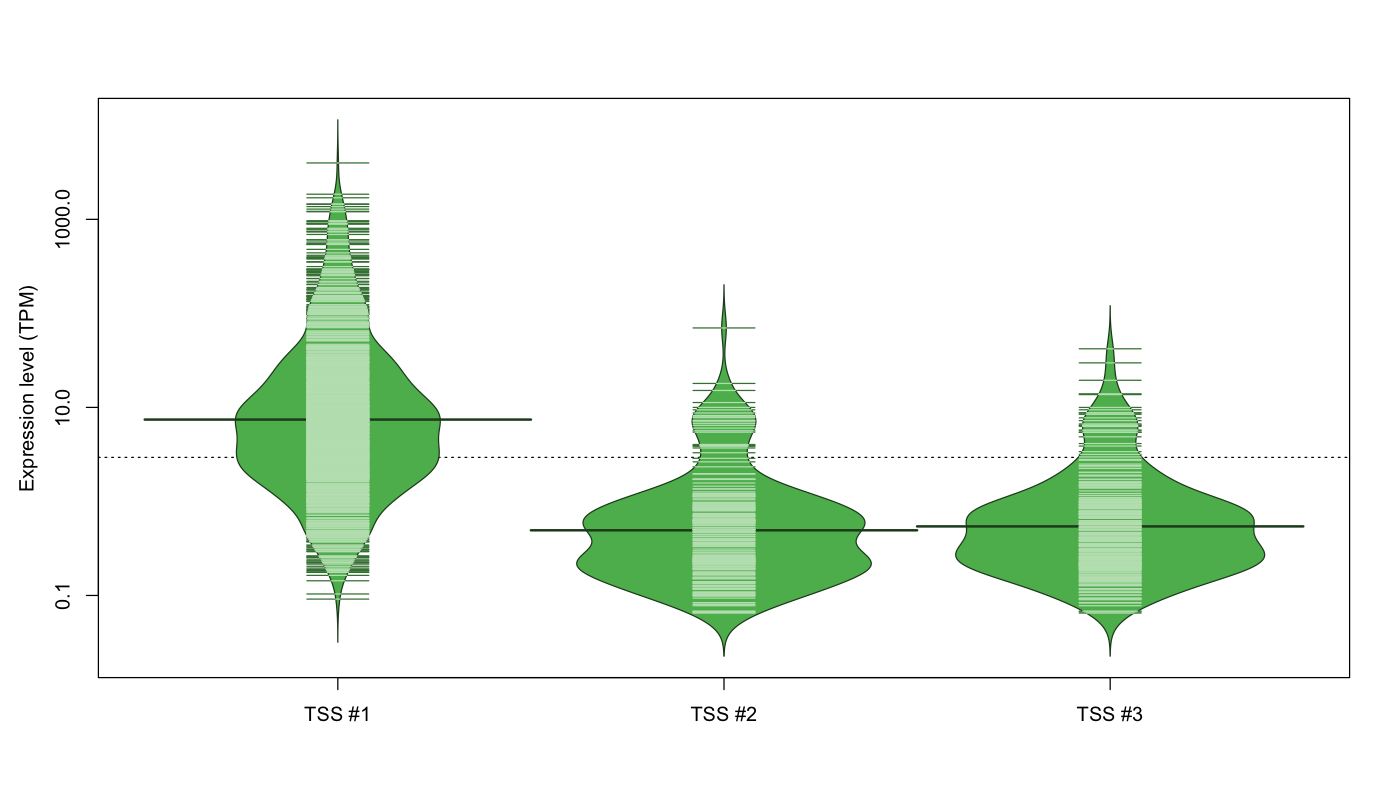
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_4382 | 147 libraries | 108 libraries | 307 libraries | 148 libraries | 362 libraries |
| TSS #2 | TSS_4383 | 851 libraries | 172 libraries | 31 libraries | 13 libraries | 5 libraries |
| TSS #3 | TSS_4384 | 799 libraries | 210 libraries | 43 libraries | 15 libraries | 5 libraries |
The graphical summary, for retro_mmus_135 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_135 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_135 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 8 parental genes, and 68 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000020554 | 3 retrocopies | |
| Homo sapiens | ENSG00000130707 | 14 retrocopies | |
| Gorilla gorilla | ENSGGOG00000010228 | 8 retrocopies | |
| Myotis lucifugus | ENSMLUG00000006066 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000076441 | 1 retrocopy |
retro_mmus_135 ,
|
| Nomascus leucogenys | ENSNLEG00000012500 | 13 retrocopies | |
| Pongo abelii | ENSPPYG00000019692 | 13 retrocopies | |
| Pan troglodytes | ENSPTRG00000021473 | 14 retrocopies |