RetrogeneDB ID: | retro_mmus_92 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 6:148354709..148355498(+) | ||
| Located in intron of: | ENSMUSG00000030306 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000063171 | |
| Aliases: | Rps4l, 1110033J19Rik, AA407494, Rps4y2 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Rps4x | ||
| Ensembl ID: | ENSMUSG00000031320 | ||
| Aliases: | Rps4x, Rps4, Rps4-1 | ||
| Description: | ribosomal protein S4, X-linked [Source:MGI Symbol;Acc:MGI:98158] |

Retrocopy-Parental alignment summary:
>retro_mmus_92
ATGGCTCGTGGTCCCAAGAAACACCTGAAGCGTGTGGCGGCCCCACGCCACTGGATGCTGGACAAACTGACCGGTGTGTT
CGCGCCCCGTCCGTCTGCCGGCCCGCACCGCCTGCGGGAGTGCCTGCCGCTCGCCATCTTCCTGAGGAACAGGCTCAAGT
ACGCCCTGACCGGCGACGAGGTGAAGAAGATCTGCATGCAGCGGCTCATTAAGGTCGACGGCAAGGTCAGAACCGATGTG
GCCTACCCAGCTGGCTTCATGGATGTCATCAGCATCGACAAGAGCGGCGAGAACTTCCGCCTCGTCTACGACACCAAGGG
CCGCTTCGCGGTACACCGCATCACGCCCGAGGAGGCCAAGTACAAGCTGTGCAAGGTGAGGAAAGTCTTCGTGGGTACCA
AGGGCATCCCGCACCTCGTCACGCACGACGCGCGTACCATCCGCTACCCTGACCCGCTCATCAAGGTCAACGACACCGTG
CAGATCTCGCTAGACAGCGGAAAAATCACCGATGCCATCAAGTTTGATACCGGCAACCTGTGCATGGTAACCGGAGGCGC
CAACCTGGGTCGCATCGGCGTCATCACCAACCGCGAGCGCCACCCCGGCTCCTTTGACGTGGTCCACGTGAAAGATGCCA
ACGGCAATGGCTTCGCTACCCGCCTCTCCAACATCTTCGTGATCGGGAAGGGCAACAAGCCGTGGATCTCCCTTCCCCGG
GGAAAAGGAATCCGCCTCACCATTGCCGAAGAGAGAGACAAGAGGCTGGCGGCCAAGCAGAGCGGGTGA
ORF - retro_mmus_92 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.49 % |
| Parental protein coverage: | 99.24 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MARGPKKHLKRVAAPKHWMLDKLTGVFAPRPSTGPHKLRECLPLIIFLRNRLKYALTGDEVKKICMQRFI |
| MARGPKKHLKRVAAP.HWMLDKLTGVFAPRPS.GPH.LRECLPL.IFLRNRLKYALTGDEVKKICMQR.I | |
| Retrocopy | MARGPKKHLKRVAAPRHWMLDKLTGVFAPRPSAGPHRLRECLPLAIFLRNRLKYALTGDEVKKICMQRLI |
| Parental | KIDGKVRTDITYPAGFMDVISIDKTGENFRLIYDTKGRFAVHRITPEEAKYKLCKVRKIFVGTKGIPHLV |
| K.DGKVRTD..YPAGFMDVISIDK.GENFRL.YDTKGRFAVHRITPEEAKYKLCKVRK.FVGTKGIPHLV | |
| Retrocopy | KVDGKVRTDVAYPAGFMDVISIDKSGENFRLVYDTKGRFAVHRITPEEAKYKLCKVRKVFVGTKGIPHLV |
| Parental | THDARTIRYPDPLIKVNDTIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHV |
| THDARTIRYPDPLIKVNDT.QI.L..GKITD.IKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHV | |
| Retrocopy | THDARTIRYPDPLIKVNDTVQISLDSGKITDAIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHV |
| Parental | KDANGNSFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQS |
| KDANGN.FATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQS | |
| Retrocopy | KDANGNGFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 8 .36 RPM | 93 .43 RPM |
| SRP007412_cerebellum | 9 .42 RPM | 46 .29 RPM |
| SRP007412_heart | 10 .12 RPM | 123 .84 RPM |
| SRP007412_kidney | 8 .01 RPM | 110 .38 RPM |
| SRP007412_liver | 8 .93 RPM | 108 .09 RPM |
| SRP007412_testis | 47 .61 RPM | 8 .42 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_92 was not detected
45 EST(s) were mapped to retro_mmus_92 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA500014 | 148355006 | 148355264 | 100 | 255 | 0 | 253 |
| AI325738 | 148354727 | 148355098 | 96.3 | 360 | 11 | 348 |
| AL117823 | 148354671 | 148354928 | 97.3 | 257 | 0 | 254 |
| AV451715 | 148354668 | 148355098 | 99.8 | 429 | 0 | 427 |
| AV462502 | 148354676 | 148355103 | 100 | 426 | 0 | 425 |
| AV463810 | 148354666 | 148355103 | 100 | 436 | 0 | 435 |
| AV464024 | 148354682 | 148355097 | 99.6 | 410 | 1 | 404 |
| AV465962 | 148354666 | 148354985 | 100 | 316 | 0 | 314 |
| AV466506 | 148354705 | 148355142 | 100 | 437 | 0 | 437 |
| AV472090 | 148354708 | 148355009 | 99 | 294 | 3 | 289 |
| AV472928 | 148354700 | 148355113 | 100 | 413 | 0 | 413 |
| AV472933 | 148354673 | 148355069 | 99.8 | 390 | 1 | 385 |
| AV482075 | 148354677 | 148355090 | 99.8 | 410 | 0 | 406 |
| AV483695 | 148354676 | 148355097 | 100 | 419 | 0 | 417 |
| AV493432 | 148354697 | 148355087 | 99.8 | 389 | 1 | 388 |
| AV494405 | 148354764 | 148354985 | 100 | 221 | 0 | 221 |
| AV496116 | 148354671 | 148355008 | 100 | 337 | 0 | 337 |
| AV498101 | 148354661 | 148355099 | 99.8 | 436 | 1 | 434 |
| AV500341 | 148354778 | 148355152 | 100 | 374 | 0 | 374 |
| AV503345 | 148354681 | 148355096 | 98.8 | 415 | 0 | 413 |
| AV506464 | 148354672 | 148355100 | 100 | 426 | 0 | 425 |
| AV506996 | 148354671 | 148355099 | 99.8 | 425 | 1 | 422 |
| AV510526 | 148354666 | 148355092 | 99.8 | 421 | 1 | 416 |
| AV511272 | 148354666 | 148355095 | 99.8 | 426 | 0 | 422 |
| AV511384 | 148354669 | 148355107 | 99.8 | 435 | 1 | 432 |
| AV515777 | 148354666 | 148355103 | 100 | 436 | 0 | 435 |
| AV516234 | 148354683 | 148355068 | 99.8 | 380 | 0 | 376 |
| AV575050 | 148354720 | 148355029 | 98.8 | 307 | 1 | 304 |
| AV581407 | 148354695 | 148355058 | 98.7 | 361 | 2 | 358 |
| AV581989 | 148354677 | 148354982 | 100 | 305 | 0 | 305 |
| AV586344 | 148354708 | 148355136 | 98.4 | 420 | 7 | 412 |
| BF012952 | 148354734 | 148355078 | 99.5 | 342 | 2 | 340 |
| BU605011 | 148354789 | 148354979 | 99 | 188 | 2 | 186 |
| BY029273 | 148354664 | 148355097 | 100 | 381 | 0 | 380 |
| BY034424 | 148354709 | 148355085 | 99.5 | 373 | 2 | 371 |
| BY036867 | 148354657 | 148355029 | 99.8 | 368 | 1 | 367 |
| BY096065 | 148354679 | 148355071 | 100 | 391 | 0 | 391 |
| BY114163 | 148354657 | 148354973 | 100 | 316 | 0 | 316 |
| BY115419 | 148354657 | 148354996 | 99.8 | 338 | 1 | 337 |
| BY321323 | 148354709 | 148355056 | 100 | 346 | 0 | 346 |
| BY328788 | 148354657 | 148354990 | 99.4 | 331 | 2 | 329 |
| CJ163072 | 148354657 | 148355047 | 100 | 390 | 0 | 390 |
| CJ164673 | 148354657 | 148355078 | 100 | 421 | 0 | 421 |
| CJ171908 | 148354686 | 148355065 | 100 | 379 | 0 | 379 |
| D76864 | 148354671 | 148354962 | 96.9 | 285 | 2 | 274 |
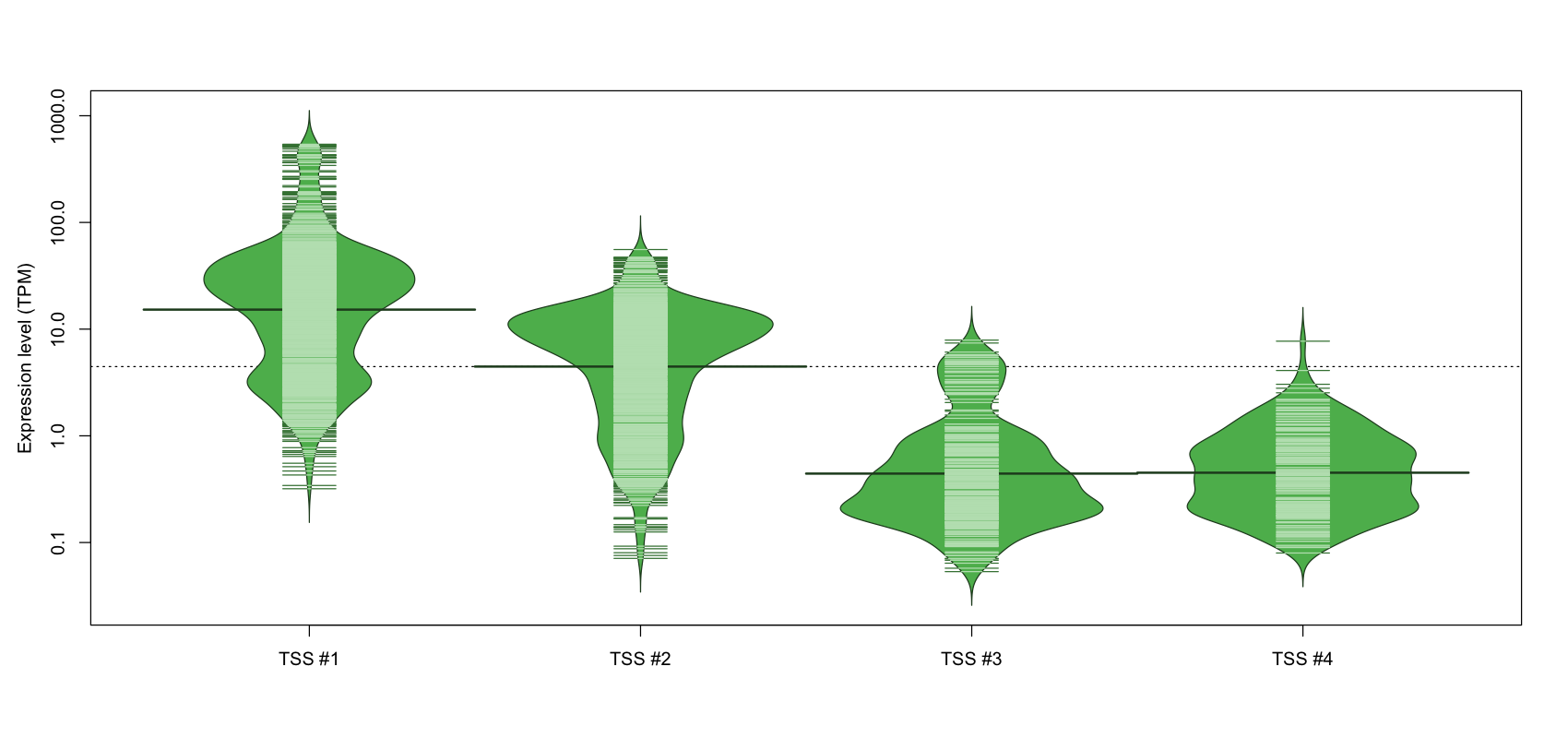
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_120399 | 131 libraries | 16 libraries | 223 libraries | 102 libraries | 600 libraries |
| TSS #2 | TSS_120400 | 224 libraries | 141 libraries | 218 libraries | 205 libraries | 284 libraries |
| TSS #3 | TSS_120401 | 777 libraries | 237 libraries | 48 libraries | 10 libraries | 0 libraries |
| TSS #4 | TSS_120402 | 864 libraries | 167 libraries | 40 libraries | 1 library | 0 libraries |
The graphical summary, for retro_mmus_92 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_92 was not experimentally validated.