RetrogeneDB ID: | retro_mmus_1608 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 17:5417511..5418513(-) | ||
| Located in intron of: | ENSMUSG00000004945 | ||
Retrocopy information | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Ldha | ||
| Ensembl ID: | ENSMUSG00000063229 | ||
| Aliases: | Ldha, Ldh1, Ldhm, l7R2 | ||
| Description: | lactate dehydrogenase A [Source:MGI Symbol;Acc:MGI:96759] |

Retrocopy-Parental alignment summary:
>retro_mmus_1608
TCCTCCATGGCGACCGTGAAGGGTGAACTTATCCAGCACATCAGCTCCGAAGAGCCCGTTCTCCACAACAAGGTCTCCAT
TGTAGGAACTGGATCAGTCGGCATGGCCTGTGCCATCGGCATCATAGCGAAAGGCTTGACTGACGAGCTCGCCCTTGTTG
ATAATAATGAAGAAAAAATGAAGGGTGAGACGATGGATCTTCAGCACGGCAGTGTCTTCATGAAAATGCCAAATATTGTT
TGCAGCAAGGATTTCCGTGTCACTGCCAACTCTGAGGTAGTGATTATCACAGCGGGTGCACGCCAAGAGAAAAATGAGAC
ACGCCTGAATTTAGTCCAGCGAAACGTAACCATCTTTAAGGCAATGGTTGCGGAAATTATCAAGCACAGCCCTCGCTGCA
AAATTATTGTTGTCTCCAATCCAGTGGATATCTTAACTTTCGTAACCTGGAAGTTGAGTGGGTTTCCCAAAAACCGTATT
ATTGGAAGCGGCTGTAACCTAGATACAGCTCGCTTCCGTTACATGCTCGGGCAGAGGCTTGGGATCCACTCTGAAAGCTG
CCACGGGTGGGTCCTTGGAGAGCATGGAGATTCAAGTGTCCCTGTGTGGAGTGGTGTGAACATCGCCGGCGTGCCGCTGA
GAGAGCTGAATTCAGCTATAGGAACAAGCAAAGACCCCGAGAAATGGGGCGATGTCCACAAAGAAGTCATTGCCAGTGCC
TATAATATTATCAAAATGAAAGGTTATACTTCCTGGGCCATTGGCCTGTCTGTAACTGATATTGCAGAGAGTATCCTGAA
GAATCTCAGGAAGACGCATCCAGTTACCACCAAAATCCAAGGTCTCTACGGAATCAAAGAGGAGGTGTTCCTCAGCGTGC
CTTGTATCCTGGGAGAGAGTGGCATATCTGATATCATAAAGGTAAAGCTGAGCCCTACAGAGGAGGCCCAGATGGTAAAG
AGCGCAGAGACTATTTGGAAAATCCAGAAAGATATTAAGTTT
ORF - retro_mmus_1608 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 67.07 % |
| Parental protein coverage: | 92.52 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | SKMATLKDQLIVNLLKEEQAPQNKITVVGVGAVGMACAISILMKDLADELALVDVMEDKLKGEMMDLQHG |
| S.MAT.K..LI.....EE....NK...VG.G.VGMACAI.I..K.L.DELALVD..E.K.KGE.MDLQHG | |
| Retrocopy | SSMATVKGELIQHISSEEPVLHNKVSIVGTGSVGMACAIGIIAKGLTDELALVDNNEEKMKGETMDLQHG |
| Parental | SLFLKTPKIVSSKDYCVTANSKLVIITAGARQQEGESRLNLVQRNVNIFKFIIPNIVKYSPHCKLLIVSN |
| S.F.K.P.IV.SKD..VTANS..VIITAGARQ...E.RLNLVQRNV.IFK.....I.K.SP.CK...VSN | |
| Retrocopy | SVFMKMPNIVCSKDFRVTANSEVVIITAGARQEKNETRLNLVQRNVTIFKAMVAEIIKHSPRCKIIVVSN |
| Parental | PVDILTYVAWKISGFPKNRVIGSGCNLDSARFRYLMGERLGVHALSCHGWVLGEHGDSSVPVWSGVNVAG |
| PVDILT.V.WK.SGFPKNR.IGSGCNLD.ARFRY..G.RLG.H..SCHGWVLGEHGDSSVPVWSGVN.AG | |
| Retrocopy | PVDILTFVTWKLSGFPKNRIIGSGCNLDTARFRYMLGQRLGIHSESCHGWVLGEHGDSSVPVWSGVNIAG |
| Parental | VSLKSLNPELGTDADKEQWKEVHKQVVDSAYEVIKLKGYTSWAIGLSVADLAESIMKNLRRVHPISTMIK |
| V.L..LN...GT..D.E.W..VHK.V..SAY..IK.KGYTSWAIGLSV.D.AESI.KNLR..HP..T.I. | |
| Retrocopy | VPLRELNSAIGTSKDPEKWGDVHKEVIASAYNIIKMKGYTSWAIGLSVTDIAESILKNLRKTHPVTTKIQ |
| Parental | GLYGINEDVFLSVPCILGQNGISDVVKVTLTPEEEARLKKSADTLWGIQKELQF |
| GLYGI.E.VFLSVPCILG..GISD..KV.L.P.EEA...KSA.T.W.IQK...F | |
| Retrocopy | GLYGIKEEVFLSVPCILGESGISDIIKVKLSPTEEAQMVKSAETIWKIQKDIKF |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .23 RPM | 162 .35 RPM |
| SRP007412_cerebellum | 0 .39 RPM | 72 .30 RPM |
| SRP007412_heart | 0 .06 RPM | 465 .65 RPM |
| SRP007412_kidney | 0 .31 RPM | 344 .67 RPM |
| SRP007412_liver | 0 .19 RPM | 394 .74 RPM |
| SRP007412_testis | 439 .09 RPM | 722 .17 RPM |
RNA Polymerase II activity may be related with retro_mmus_1608 in 2 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001YJZ | POLR2A | 17:5408855..5419001 |
| ENCFF001YKA | POLR2A | 17:5419049..5419312 |
| ENCFF001YKA | POLR2A | 17:5418449..5418670 |
No EST(s) were mapped for retro_mmus_1608 retrocopy.
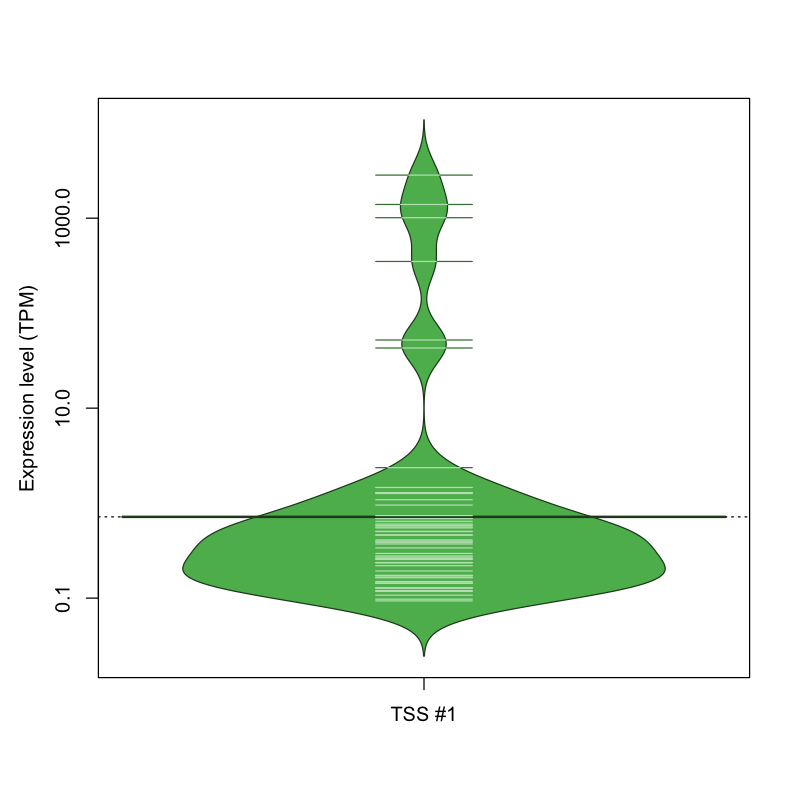
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_55298 | 1018 libraries | 42 libraries | 6 libraries | 0 libraries | 6 libraries |
The graphical summary, for retro_mmus_1608 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | GCAGAAGAGTGAGTACAACC (20 nt long) |
| Reverse primer: | CACACTTATGAAAGTTAGCCG (21 nt long) |
| Annealing temperature: | 55 °C |
| (Expected) product size: | 1362 |
| Electrophoresis gel image: | 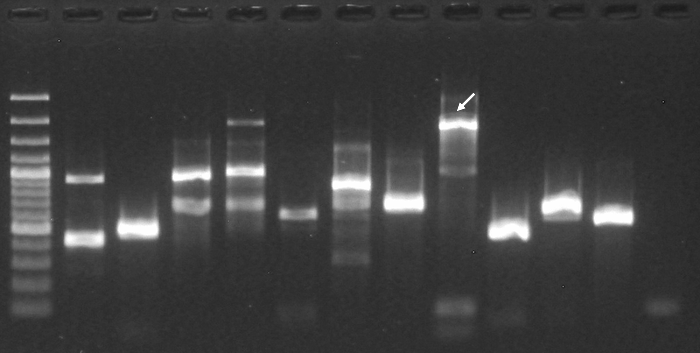 |
| Additional comment: | Pfu DNA Polymerase; pooled mouse cDNA; 40 cycles of PCR |
Retrocopy orthology:
Retrocopy retro_mmus_1608 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_578 |